A selection of publications authored by the BRENDA team.
Schomburg I., Jeske L., Ulbrich M., Placzek S., Chang A., Schomburg D. (2017) The BRENDA enzyme information system – From a database to an expert system. J. Biotechnol. 261, 194-206. PubMed
Placzek S., Schomburg I., Chang A., Jeske L., Ulbrich M., Tillack J., Schomburg D. (2017) BRENDA in 2017: new perspectives and new tools in BRENDA. Nucleic Acids Res. 45, D380-D388. Oxford
Chang A., Schomburg I., Placzek S., Jeske L., Ulbrich M., Xiao M., Sensen C.W., Schomburg D. (2015) BRENDA in 2015: exciting developments in its 25th year of existence. Nucleic Acids Res. 43, D439-D446. Oxford
Schomburg I., Chang A., Placzek S., Söhngen C., Rother M., Lang M., Munaretto C., Ulas S., Stelzer M., Grote A. Scheer M., Schomburg D. (2013) BRENDA in 2013: integrated reactions, kinetic data, enzyme function data, improved disease classification: new options and contents in BRENDA. Nucleic Acids Res. 41, 764-772. PubMed
Gremse M., Chang A., Schomburg I., Grote A., Scheer M., Ebeling C., Schomburg D.(2011) The BRENDA Tissue Ontology (BTO): the first all-integrating ontology of all organisms for enzyme sources. Nucleic Acids Res. 39, D507-D513. PubMed
Barthelmes J., Ebeling C., Chang A., Schomburg I., Schomburg D. (2007) BRENDA, AMENDA and FRENDA: the enzyme information system in 2007. Nucleic Acids Res. 35, D511-D514. PubMed
Schomburg I., Chang A., Hofmann O., Ebeling C., Ehrentreich F., Schomburg D., (2002) BRENDA: a resource for enzyme data and metabolic information. Trends Biochem. Sci. 27, 54-56. PubMed
Schomburg, I., Chang, A., Schomburg, D., (2002) BRENDA, enzyme data and metabolic information. Nucleic Acids Res. 30, 47-49. PubMed
Schomburg, D., Schomburg, I. (2001) Springer Handbook of Enzymes. 2nd Ed. Springer, Heidelberg. Springer
Schomburg, I., Hofmann, O., Baensch, C., Chang, A., Schomburg, D., (2000) Enzyme data and metabolic information: BRENDA, a resource for research in biology, biochemistry, and medicine. Gene Funct. Dis. 3-4, 109-18.







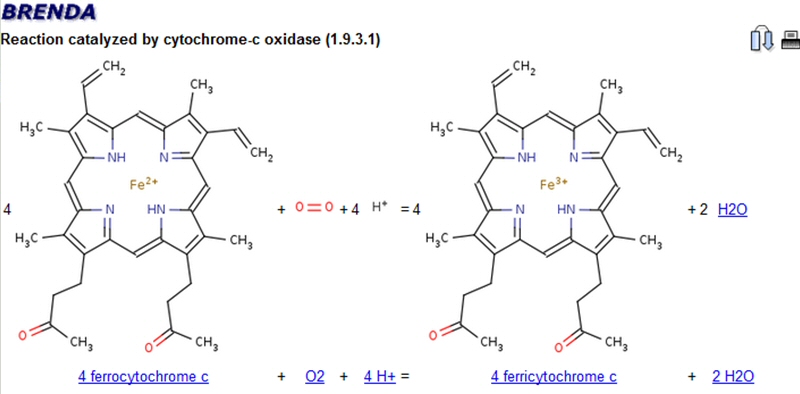
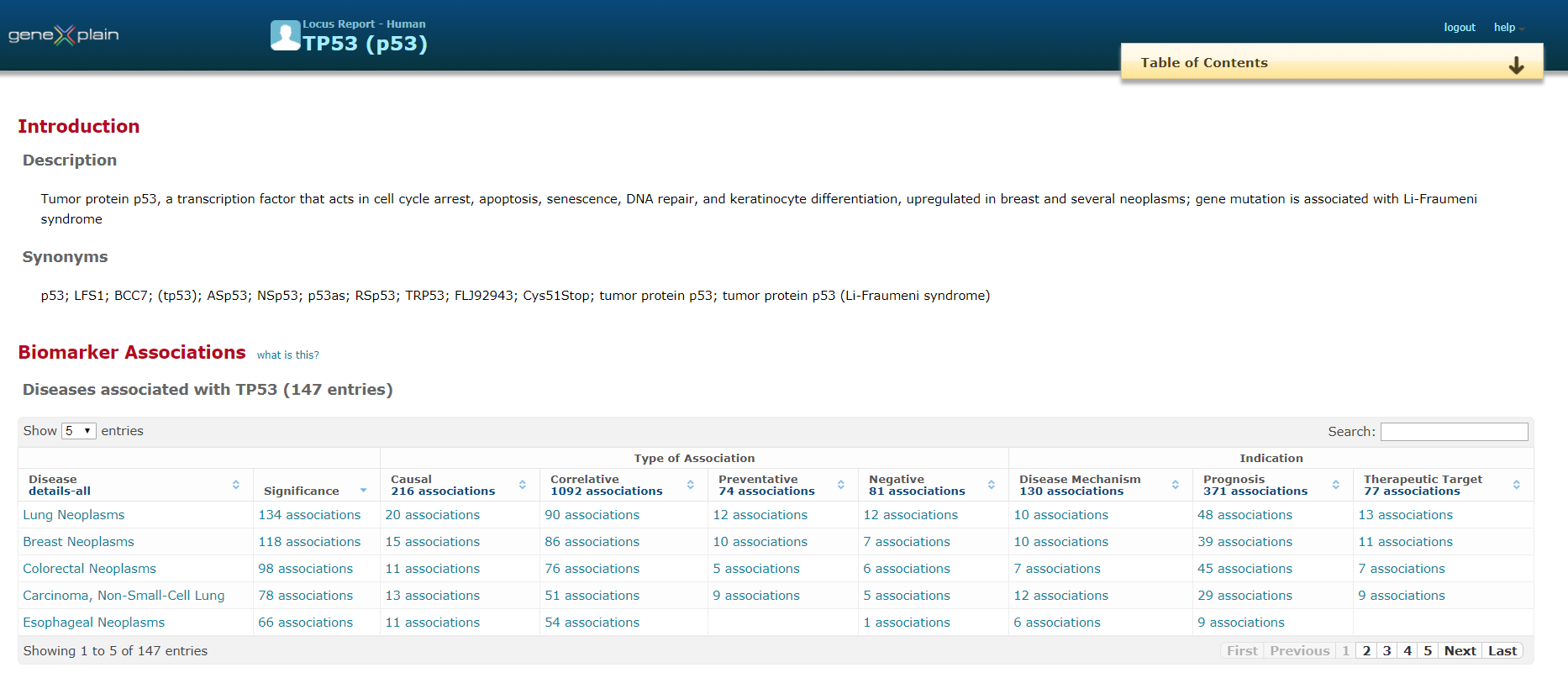
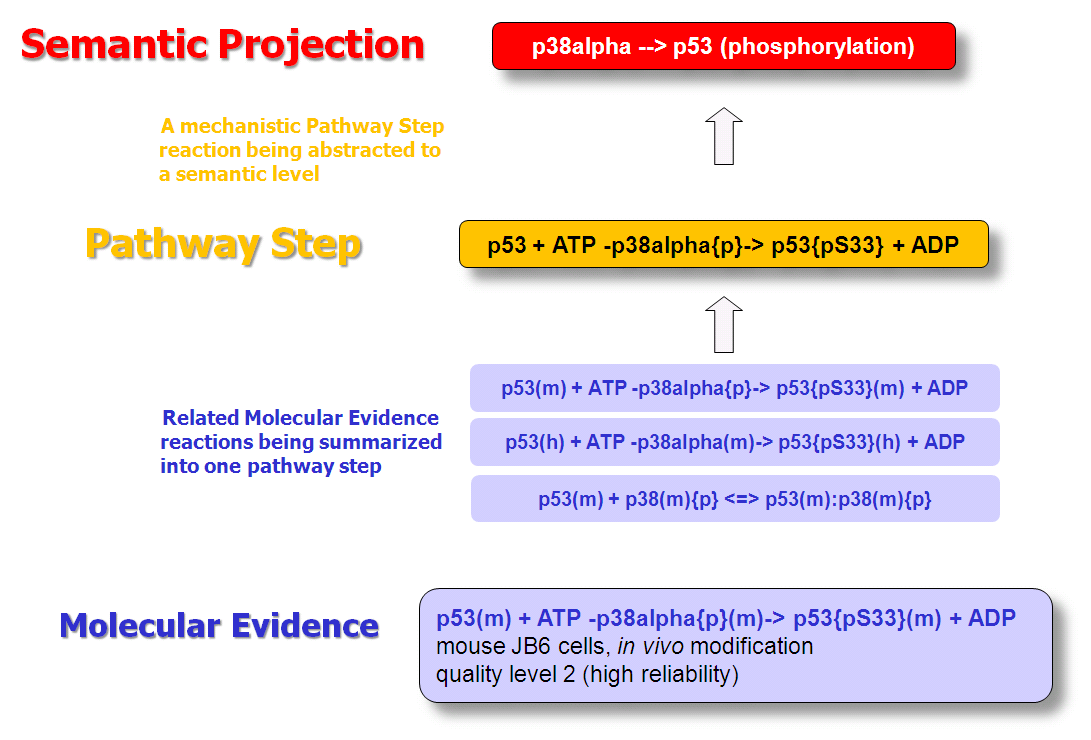
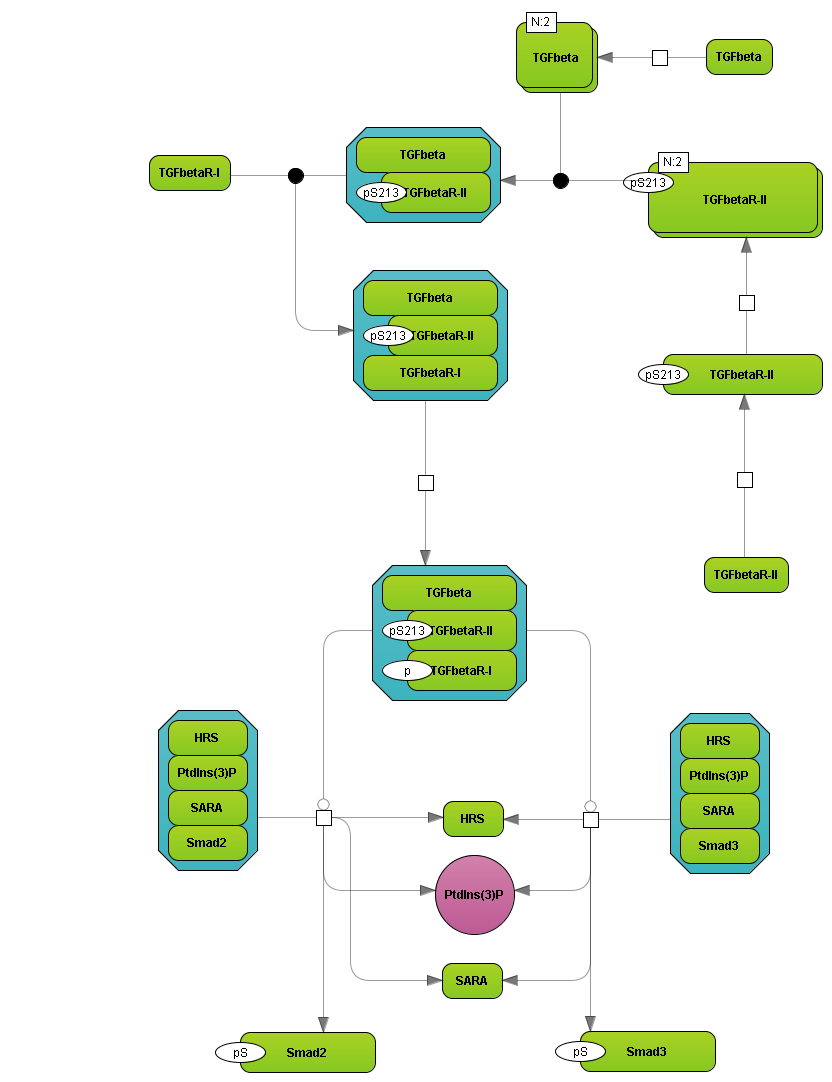
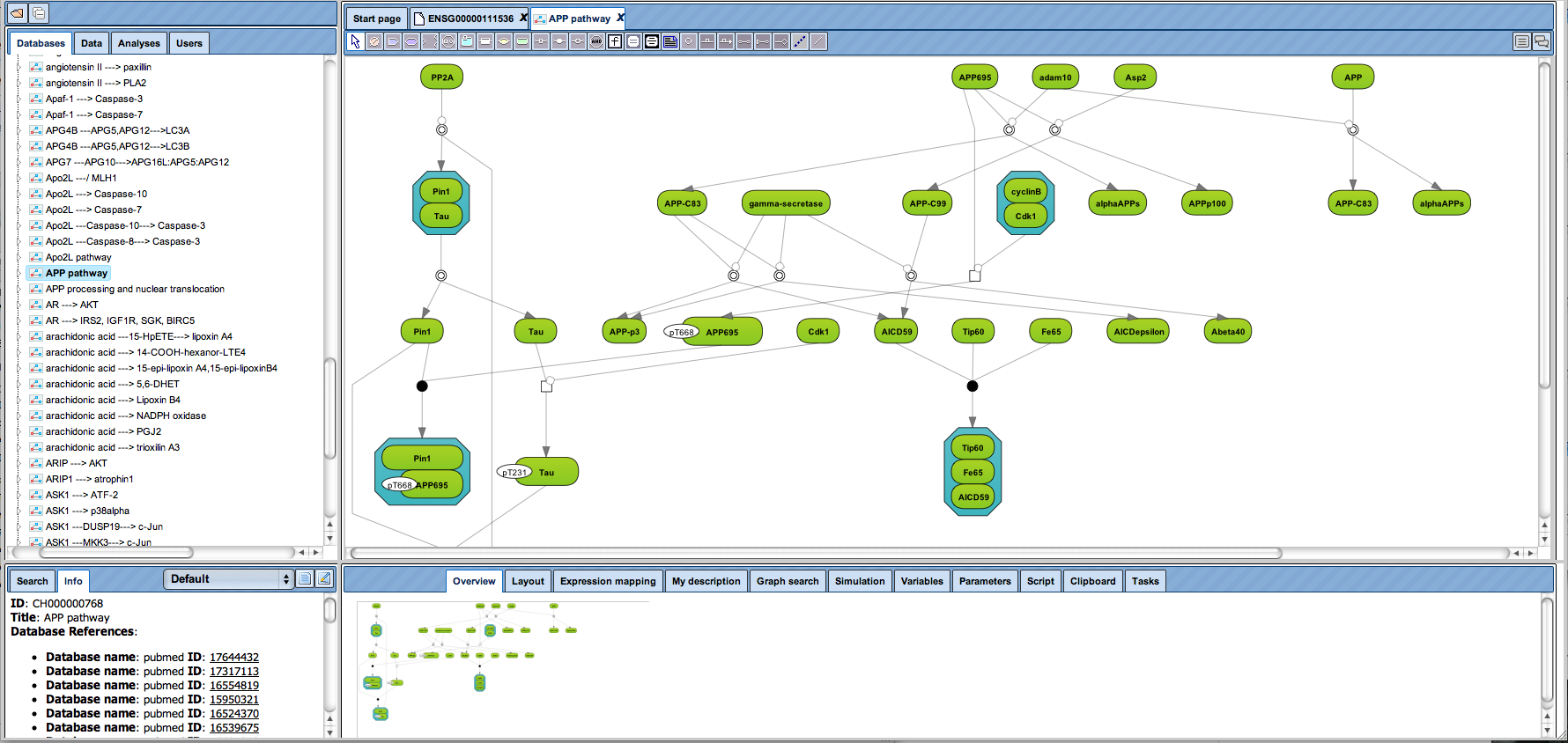
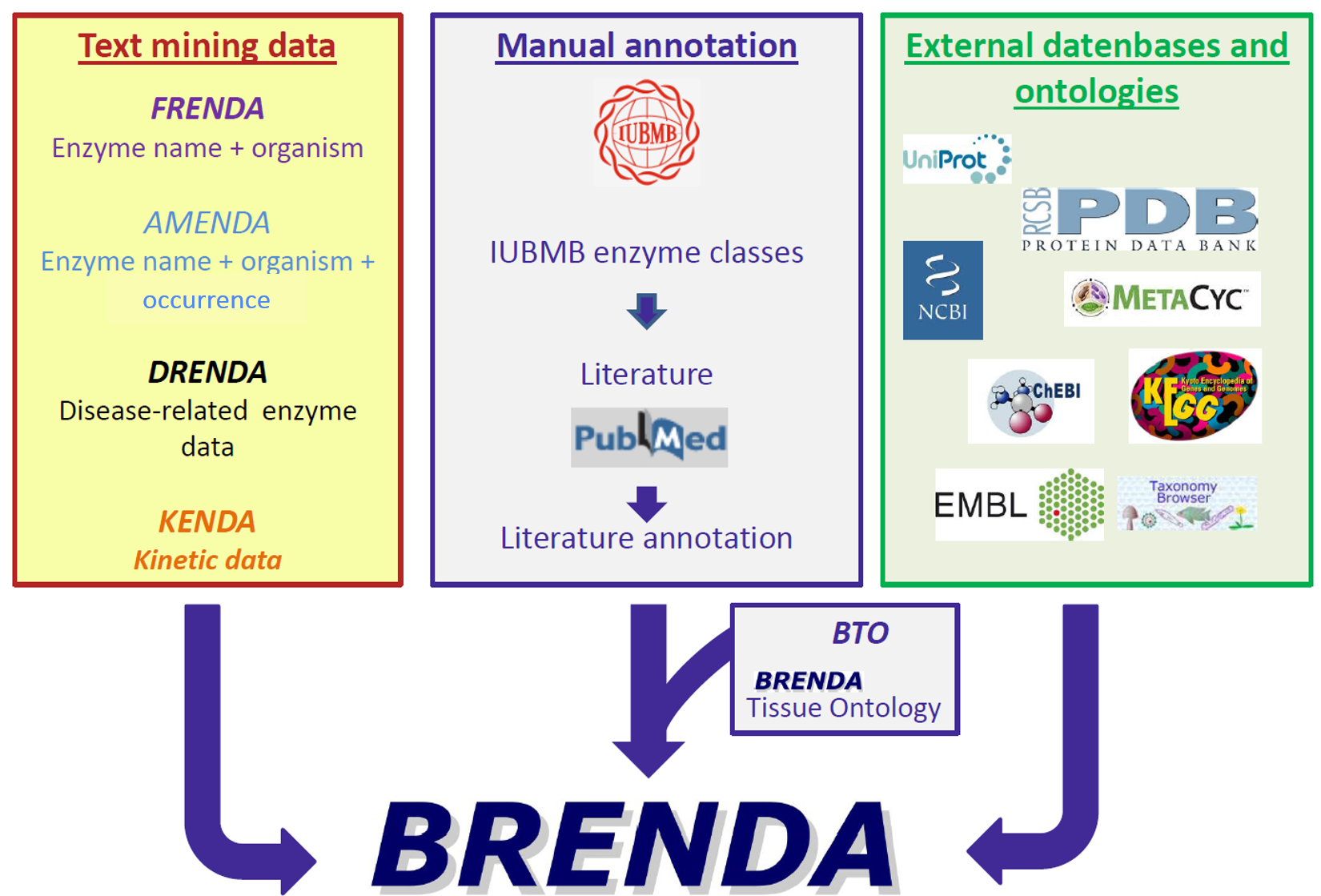 Organization of BRENDA® contents
Organization of BRENDA® contents






 Subscribe to our channel
Subscribe to our channel