geneXplain platform API
The geneXplain platform provides a comprehensive environment to analyze biomedical and biological data. Its functionality includes, among other things, data storage, data management, data sharing, in-built bioinformatics and systems biology analysis tools, ability to build and run analysis pipelines and workflows, build and visualize molecular network models, or develop quantitative models and perform simulations. More details about the platform can be found on the platform product page and in the corresponding research articles.
The geneXplain platform can be used through a graphical web interface or through APIs (Application Programming Interfaces) that have been implemented in the languages Java and R. Besides being integrated as a software library, the genexplain-api package provides several commandline utilities. The exec tool allows to configure and run remote analyses using JSON input files. The JSON interface is described in detail in the genexplain-api documentation.
The following figure sketches some of the tasks that can be carried out with the platform APIs:
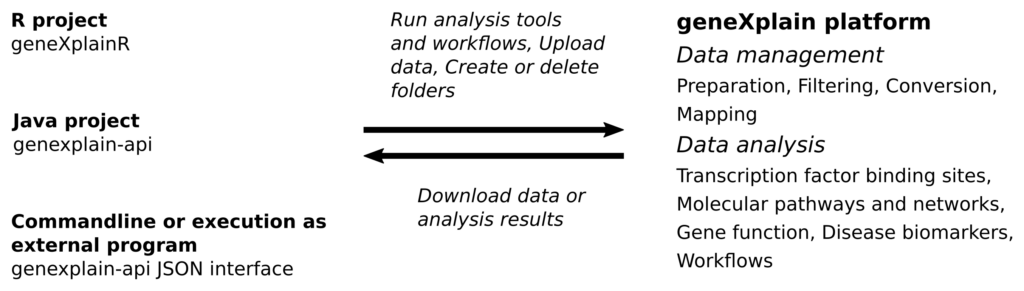
All analysis and utility tools provided by the geneXplain platform as well as integrated Galaxy tools and workflows can be executed using API functions. In addition, there are methods to manage, organize and download research data and analysis results. Notably, analysis jobs can run asynchronously so that you are not required to wait for long analysis tasks.
You are welcome to view the geneXplain platform API tutorial covering both: the geneXplain platform R and the geneXplain platform Java APIs, as well as the command line interface, here.
geneXplain platform R API (geneXplainR)
GeneXplainR package provides an interface for the easier integration of the geneXplain platform functionalities into the R pipelines. Combine the wide range of data, tools, and workflows of the geneXplain platform with the resources available in R language to perform your bioinformatic analysis.
The geneXplainR package provides an R client for the geneXplain platform, geneXplainR is based on and extends the rbiouml package. A goal of this project is to add functionality that helps to make building R pipelines that use the geneXplain platform easier.
Watch this video demonstrating the example of executing the “Identify enriched motif in promoters (TRANSFAC)” workflow of the geneXplain platform using the R API:
Check out the record of Coffee break with TRANSFAC webinar devoted to the geneXplain platform API:
geneXplain platform Java API
The geneXplain Java API allows integration of geneXplain platform functionalities into Java programs and provides a JSON interface to configure and execute individual analysis tasks as well as complex workflows. The geneXplain platform Java API can be used to write Java programs that can invoke functionality of the platform through its web interface, e.g. for import and export of biological data, or to submit and monitor analysis jobs. Furthermore, platform tasks can be specified in JSON format and submitted using the executable JAR file. Though not endowed with all possiblities of a programming language, the JSON interface facilitates, among other things, definition of templates, branch points, or nesting of tasks, so that you can build complex workflows from reusable components. It is intended to be applied as part of a dynamic and polyglot analysis environment that utilizes diverse programming languages and resources.
Watch thIs introductory tutorial on geneXplain platform Java API that shows how to install the API software and run the basic commands:
