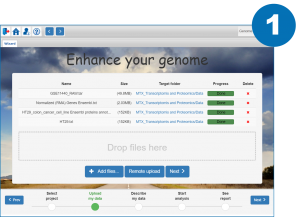
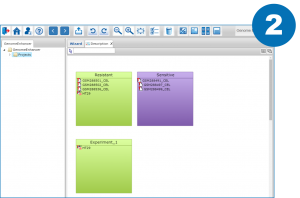
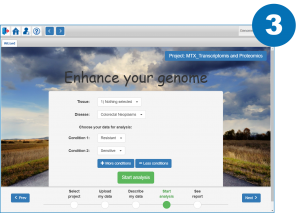
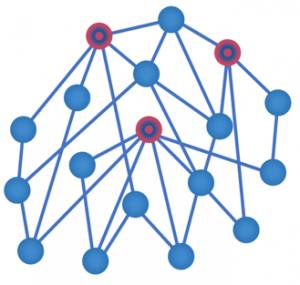
References
- Lloyd, Katie, et al. “Using systems medicine to identify a therapeutic agent with potential for repurposing in Inflammatory Bowel Disease.” Disease models & mechanisms (2020).
- Kel, Alexander, et al. “Walking pathways with positive feedback loops reveal DNA methylation biomarkers of colorectal cancer.” BMC bioinformatics 20.4 (2019): 119.
- Kolpakov, Fedor, et al. “BioUML: an integrated environment for systems biology and collaborative analysis of biomedical data.” Nucleic acids research 47.W1 (2019): W225-W233.
- Boyarskikh, Ulyana, et al. “Computational master-regulator search reveals mTOR and PI3K pathways responsible for low sensitivity of NCI-H292 and A427 lung cancer cell lines to cytotoxic action of p53 activator Nutlin-3.” BMC medical genomics 11.1 (2018): 12.
- Boyarskikh, U. A., et al. “Master-regulators driving resistance of non-small cell lung cancer cells to p53 reactivator Nutlin-3.” Virtual Biology 4 (2017): 1-31.
Disclaimer
The results of Genome Enhancer analysis, contained in any of the reports produced by this pipeline, are intended for research use only and should not be used for medical or professional advice. GeneXplain GmbH makes no guarantee of the comprehensiveness, reliability or accuracy of the information contained in the reports generated by Genome Enhancer.
Decisions regarding care and treatment of patients should be fully made by attending doctors. The predicted chemical compounds listed in the reports are given only for doctor’s consideration and they cannot be treated as prescribed medication. It is the physician’s responsibility to independently decide whether any, none or all of the predicted compounds can be used solely or in combination for patient treatment purposes, taking into account all applicable information regarding FDA prescribing recommendations for any therapeutic and the patient’s condition, including, but not limited to, the patient’s and family’s medical history, physical examinations, information from various diagnostic tests, and patient preferences in accordance with the current standard of care. Whether or not a particular patient will benefit from a selected therapy is based on many factors and can vary significantly.
The compounds predicted to be active against the identified drug targets in the reports are not guaranteed to be active against any particular patient’s condition. GeneXplain GmbH does not give any assurances or guarantees regarding the treatment information and conclusions given in the reports. There is no guarantee that any third party will provide a refund for any of the treatment decisions made based on these results. None of the listed compounds was checked by Genome Enhancer for adverse side-effects or even toxic effects.
The analysis reports contain information about chemical drug compounds, clinical trials and disease biomarkers retrieved from the HumanPSD™ database of gene-disease assignments maintained and exclusively distributed worldwide by geneXplain GmbH. The information contained in this database is collected from scientific literature and public clinical trials resources. It is updated to the best of geneXplain’s knowledge however we do not guarantee completeness and reliability of this information leaving the final checkup and consideration of the predicted therapies to the medical doctor. In all cases, the end user (including researchers and medical doctors) accepts full responsibility for all risks associated with using of information, contained in the reports generated by Genome Enhancer.
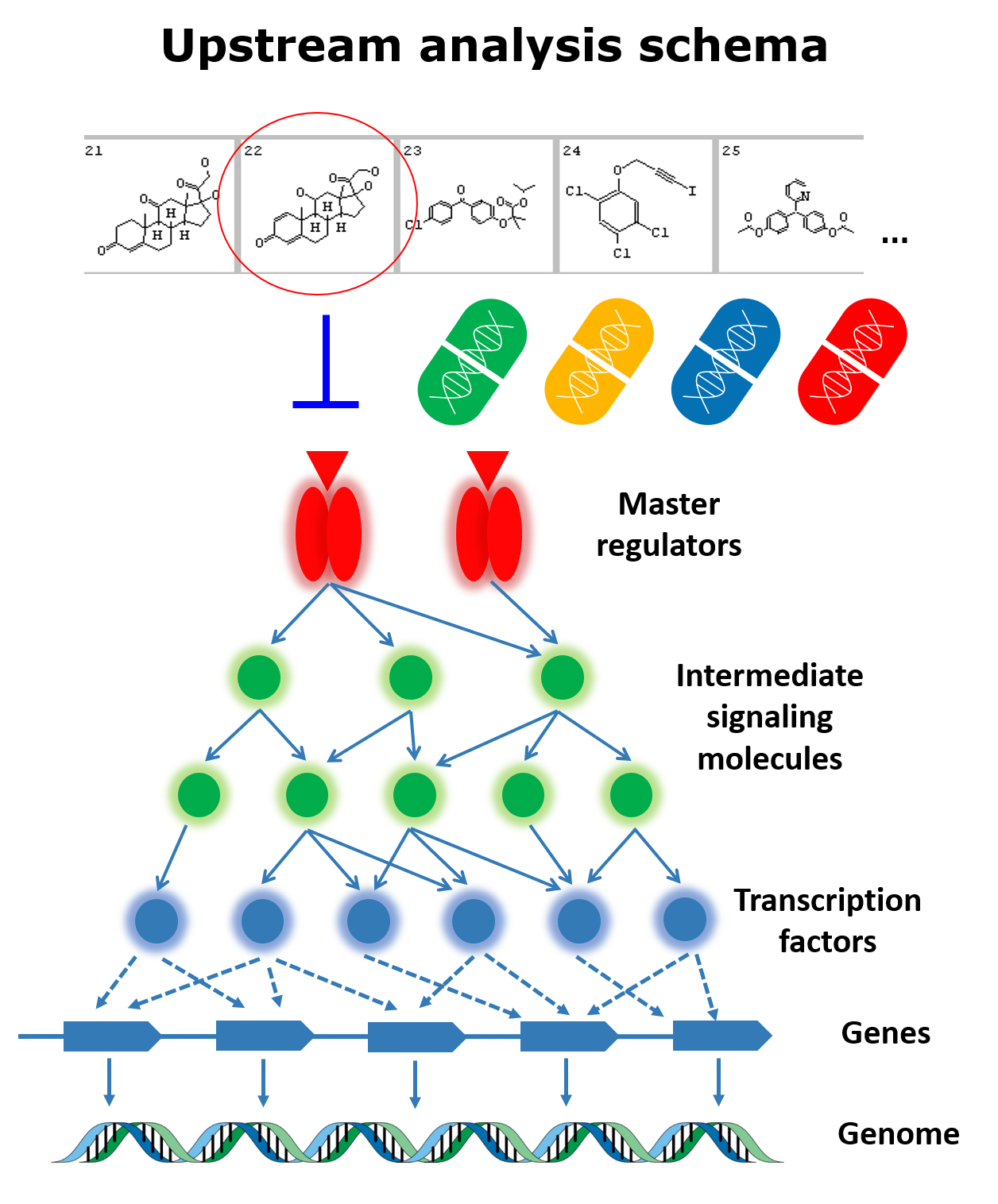
The scientific analysis underlying the Genome Enhancer reports employs a complex analysis pipeline which uses geneXplain’s proprietary Upstream Analysis approach, integrated with TRANSFAC® and TRANSPATH® databases maintained and exclusively distributed worldwide by geneXplain GmbH. The pipeline and the databases are updated to the best of geneXplain’s knowledge and belief, however, geneXplain GmbH shall not give a warranty as to the characteristics or to the content and any of the results produced by Genome Enhancer. Moreover, any warranty concerning the completeness, up-to-dateness, correctness and usability of Genome Enhancer information and results produced by it, shall be excluded.
The results produced by Genome Enhancer, including the analysis reports, severely depend on the quality of input data used for the analysis. It is the responsibility of Genome Enhancer users to check the input data quality and parameters used for running the Genome Enhancer pipeline.
Note that the text given in the reports is not unique and can be fully or partially repeated in other Genome Enhancer analysis reports, including reports of other users. This should be considered when publishing any results or excerpts from the reports. This restriction refers only to the general description of analysis methods used for generating the reports. All data and graphics referring to the concrete set of input data, including lists of mutated genes, differentially expressed genes/proteins/metabolites, functional classifications, identified transcription factors and master regulators, constructed molecular networks, lists of chemical compounds and reconstructed model of molecular mechanisms of the studied pathology are unique in respect to the used input data set and Genome Enhancer pipeline parameters used for the current run.