Last chance to get GE free trial

The New Era of Precision Medicine
Genome Enhancer free trials were stopped on January 20th 2023. You can request a license here.
Genome Enhancer is a fully automatized bioinformatics pipeline for human omics data analysis. It takes genomics, transcriptomics, proteomics, epigenomics, and metabolomics data in any combinations as input and reconstructs the molecular mechanism of the studied pathology based on the Upstream Analysis approach. As a result, Genome Enhancer generates a report on the performed analysis, which contains information about the target genes that were identified as the ones characterising the studied pathology; the transcription factors and their combinations regulating those genes; the master-regulators identified in pathway analysis as key regulators of the studied pathology; and promising drug targets and therapeutic agents acting on them in the studied case.
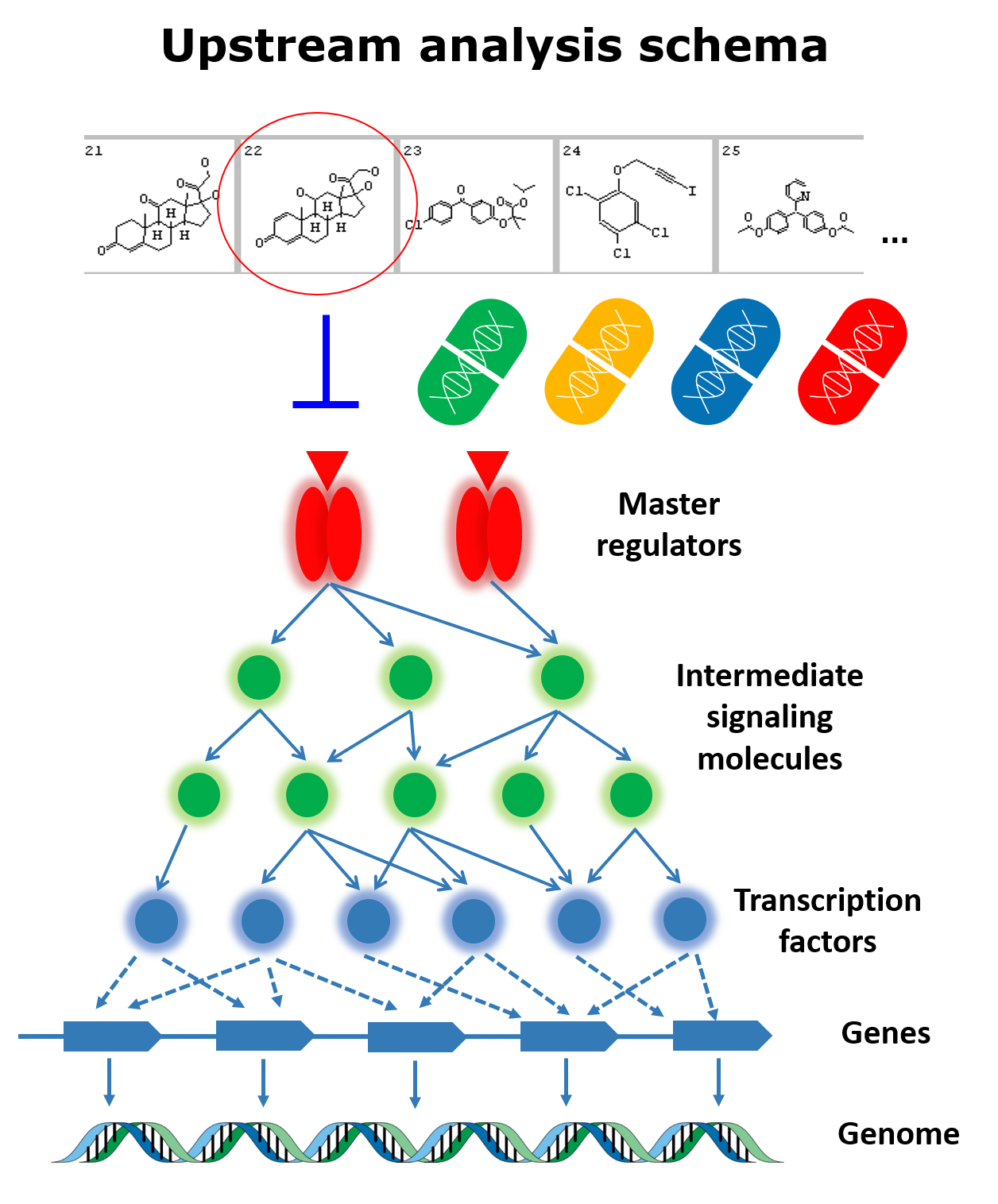
In its complex algorithm, Genome Enhancer refers to the best-in-class databases of transcription factor binding sites (TRANSFAC), intracellular reactions and pathways (TRANSPATH) and gene-drug-disease assignments (HumanPSD) maintained and exclusively distributed worldwide by geneXplain GmbH. Moreover, the backbone of Genome Enhancer is a comprehensive bioinformatics platform (geneXplain platform), which can serve as a unified solution for biomedical studies. The workflow behind the Genome Enhancer analysis is generated on the fly depending on the input omics data submitted for the run. In case of your interest, you can explore the algorithm behind the Genome Enhancer analysis here.
Genome Enhancer was first released in the year 2018 and since then was actively developed and maintained by geneXplain GmbH becoming a true game changer in the area of patient omics data analysis. Since recent releases Genome Enhancer received extended functionality for the analysis of cancer-related pathologies, including the generation of Molecular Tumor Board report. Its active development was well met by the biomedical community and we are proud to conclude that Genome Enhancer has a vast number of proven applications, including cancer, neurodegenerative diseases, infectious diseases, diabetes and metabolic diseases, and hypertension. What was previously available to patients only via long-running clinical trials, is now possible within just several hours with Genome Enhancer, which is of crucial importance for complicated clinical cases with no obvious prospective treatment schemas available. Personalized approved and off-label treatments are suggested by the system based on the molecular drug targets identified in the studied pathology for each particular clinical case, or a group of cases when cohort studies are performed.
Further info
To our delight it appeared that not only clinical doctors expressed their interest towards Genome Enhancer. A lot of biomedical researchers and students worldwide have made great use of Genome Enhancer in their studies.
The broad range of different results produced by Genome Enhancer at the intermediate steps of its analysis allow users to identify such important things as:
- Lists of target genes (genes carrying sequence variations / differentially expressed genes / genes corresponding to the differentially expressed proteins / genes with differential methylation status / genes encoding enzymes metabolizing given metabolites – depending on the input omics data submitted for the analysis)
- Functional classification of the identified target genes, including:
- GO (biological process)
- TRANSPATH® Pathways
- HumanPSD(TM) diseases
- Lists of transcription factors that potentially regulate the target genes
- Lists of master regulators that are predicted to be governing the regulation of the target genes
- Lists of prospective drug targets that can be affected by known drugs or drugs undergoing clinical trials
- Lists of prospective treatments matching the predicted targets
All this in a fully automatized pipeline within just several hours of online analysis!
Genome Enhancer works with raw or pre-processed human genomics, transcriptomics, epigenomics, proteomics, and metabolomics data coming in the following formats:
Transcriptomics (RNA-seq, microarrays)
*.txt, *.csv, *.xls (table with gene identifiers)
*.CEL (affymetrix)
*.txt (special agilent format)
*.txt (special illumina format)
*.fastq
Epigenomics (ChIP-seq)
*.fastq
*.bam (hg38 only)
*.bed (hg38 only)
*.txt (table with illumina methylation probe ids, cg*)
Genomics
*.vcf
*.txt, *.csv, *.xls (table data with SNP identifiers, rs*), *.tsv
*.fastq
Proteomics
*.txt, *.csv, *.xls (table with protein identifiers)
Metabolomics
*.txt, *.csv, *.xls (table with the list of metabolites from chebi database, e.g. CHEBI:57316)
Files of one data format can be uploaded in a .zip archive
The automatically generated analysis report is structured in a form of scientific publication, explaining all the methods used and the results obtained.
Here is an example report that was generated from raw transcriptomics FASTQ files retrieved from GSE32424 where Esophageal Squamous Cell Carcinoma was studied:
Esophageal Squamous Cell Carcinoma (GSE32424) — Transcriptomics, FASTQ
Other demo reports of Genome Enhancer are presented below:
Colorectal Cancer (Personalized patient data) — Genomics, VCF
MTB (Molecular Tumor Board) report example for colorectal cancer patient — Genomics, VCF
Esophageal Squamous Cell Carcinoma (GSE32424) — Transcriptomics, FASTQ
IFN-alpha induction (GSE31193) — Transcriptomics, LogFC Table
Lung cancer, treatment by TGF (ST000010) — Metabolomics, Table
Ovarian cancer, cisplatin-resistance (GSE15709) — Transcriptomics + Epigenomics, CEL + BED
SNP associated with Diabetes Mellitus — Genomics, SNP list
Non-Small Cell Lung Carcinoma (NCI-H1975) — Genomics, VCF
Hypertension (GSE157131) — Epigenomics, cg lists
Clinical interpretation of Genome Enhancer results is discussed in this video demonstrating how sensitivity towards VEGFA-targeted therapy was predicted by Genome Enhancer for three colorectal cancer patients.
Genome Enhancer also has its own video channel ran by the CEO of geneXplain GmbH Dr. Alexander Kel. In his videos, Alexander shows how various omics data can be analyzed in Genome Enhancer and interpreted for further usage in your studies.
End of Genome Enhancer free trials
Genome Enhancer free trials were completely stopped on January 20th, 2023.
To use Genome Enhancer, request a license for it on this page.
Disclaimer
The results of Genome Enhancer analysis, contained in any of the reports produced by this pipeline, are intended for research use only and should not be used for medical or professional advice. GeneXplain GmbH makes no guarantee of the comprehensiveness, reliability or accuracy of the information contained in the reports generated by Genome Enhancer.
Decisions regarding care and treatment of patients should be fully made by attending doctors. The predicted chemical compounds listed in the reports are given only for doctor’s consideration and they cannot be treated as prescribed medication. It is the physician’s responsibility to independently decide whether any, none or all of the predicted compounds can be used solely or in combination for patient treatment purposes, taking into account all applicable information regarding FDA prescribing recommendations for any therapeutic and the patient’s condition, including, but not limited to, the patient’s and family’s medical history, physical examinations, information from various diagnostic tests, and patient preferences in accordance with the current standard of care. Whether or not a particular patient will benefit from a selected therapy is based on many factors and can vary significantly.
The compounds predicted to be active against the identified drug targets in the reports are not guaranteed to be active against any particular patient’s condition. GeneXplain GmbH does not give any assurances or guarantees regarding the treatment information and conclusions given in the reports. There is no guarantee that any third party will provide a refund for any of the treatment decisions made based on these results. None of the listed compounds was checked by Genome Enhancer for adverse side-effects or even toxic effects.
The analysis reports contain information about chemical drug compounds, clinical trials and disease biomarkers retrieved from the HumanPSD™ database of gene-disease assignments maintained and exclusively distributed worldwide by geneXplain GmbH. The information contained in this database is collected from scientific literature and public clinical trials resources. It is updated to the best of geneXplain’s knowledge however we do not guarantee completeness and reliability of this information leaving the final checkup and consideration of the predicted therapies to the medical doctor. In all cases, the end user (including researchers and medical doctors) accepts full responsibility for all risks associated with using of information, contained in the reports generated by Genome Enhancer.
The scientific analysis underlying the Genome Enhancer reports employs a complex analysis pipeline which uses geneXplain’s proprietary Upstream Analysis approach, integrated with TRANSFAC® and TRANSPATH® databases maintained and exclusively distributed worldwide by geneXplain GmbH. The pipeline and the databases are updated to the best of geneXplain’s knowledge and belief, however, geneXplain GmbH shall not give a warranty as to the characteristics or to the content and any of the results produced by Genome Enhancer. Moreover, any warranty concerning the completeness, up-to-dateness, correctness and usability of Genome Enhancer information and results produced by it, shall be excluded.
The results produced by Genome Enhancer, including the analysis reports, severely depend on the quality of input data used for the analysis. It is the responsibility of Genome Enhancer users to check the input data quality and parameters used for running the Genome Enhancer pipeline.
Note that the text given in the reports is not unique and can be fully or partially repeated in other Genome Enhancer analysis reports, including reports of other users. This should be considered when publishing any results or excerpts from the reports. This restriction refers only to the general description of analysis methods used for generating the reports. All data and graphics referring to the concrete set of input data, including lists of mutated genes, differentially expressed genes/proteins/metabolites, functional classifications, identified transcription factors and master regulators, constructed molecular networks, lists of chemical compounds and reconstructed model of molecular mechanisms of the studied pathology are unique in respect to the used input data set and Genome Enhancer pipeline parameters used for the current run.
