TRANSFAC Professional vs TRANSFAC Public
TRANSFAC® Professional vs TRANSFAC® Public
What to pay for and is it worth it?
Sometimes users ask us: and what is the major difference between the free TRANSFAC® Public version that we can get at your public gene-regulation.com web site and the TRANSFAC® Professional solution that you charge for?
The short answer is the size of the matrix library behind the database and the quality of the output predictions that you can get using the free version compared to the professional one.
The longer answer would contain description of tools and search options integrated into the TRANSFAC® Professional, as well as the official database statistics one can explore and realise what a mass of data and knowledge stands behind our best-in-class manually curated gene regulation database.
The Professional version content is more than ten years ahead of the public release and includes data and analysis capabilities not available in the public version:
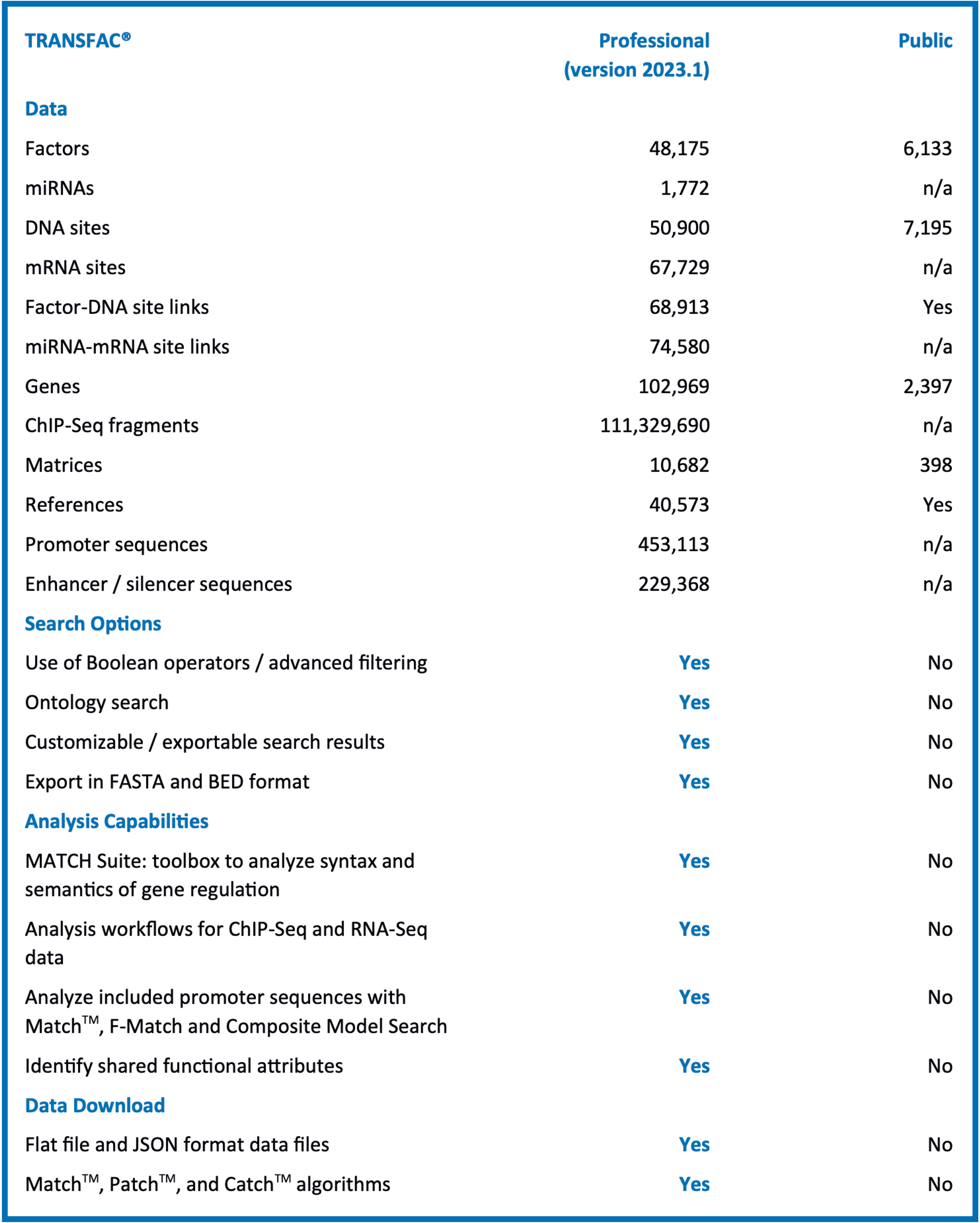
The Professional version includes advanced search options, featuring:
- Use of Boolean operators / advanced filtering
- Ontology search
- Customizable / exportable search results
- Export in FASTA and BED format
As well as extended analysis capabilities, including:
- MATCH Suite: toolbox to analyze syntax and semantics of gene regulation (human)
- Analysis workflows for ChIP-Seq and RNA-Seq data
- Analyze included promoter sequences with MatchTM, F-Match and Composite Model Search
- Identify shared functional attributes
And data Download options (Flat file download release):
- Flat file and JSON format data files
- MatchTM, PatchTM, and CatchTM algorithms
Since the analysis results are always the most convincing argument, we would like to show you the difference in the outputs that one could get while comparing TRANSFAC® Public with TRANSFAC® Professional on the same input sequence:
Transcription factor binding site prediction with MATCHTM:
A: Analysis result with the MATCHTM tool in TRANSFAC® Professional:
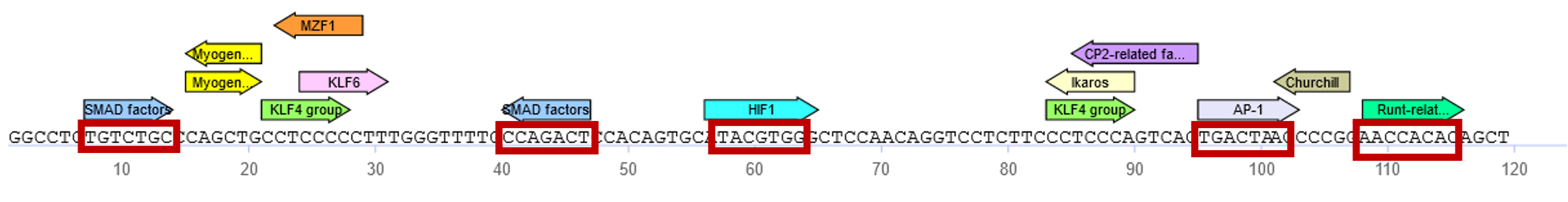

Analysis of 120 nucleotides of the human VEGFA promoter with the MATCHTM tool in TRANSFAC® Professional predicts fourteen sites for eleven different transcription factors (Fig. A). At least for four of these factors, SMAD, HIF1, AP-1 and Runx2 (predicted binding sites framed in red in the figure above), supporting evidence has been published as referenced in the TRANSFAC® professional database: “Our findings indicate that Smad directly binds to Vegfa chromatin and represses Vegfa transcriptional activity.” (PMID: 23863481) “Site-directed mutational analysis of the VEGF gene promoter revealed that an HIF-1 binding site (HBS) and its downstream HIF-1 ancillary sequence (HAS) within the HRE are required as cis-elements for the transcriptional activation of VEGF by either hypoxia or nitric oxide (NO).” (PMID: 11056166) “A gel mobility shift assay showed that the inhibitory effect of CSA was associated with decreased AP-1 binding activity to the VEGF promoter, in a cAMP-dependent manner.” (PMID: 12115224) “Physical and functional interactions between Runx2 and HIF-1α induce vascular endothelial growth factor gene expression.” (PMID: 21793044)
With MATCHTM on the public web site, at the same cut-offs, only one of these sites (AP-1) is detected (Fig. B).
Parameters:
Analyzed sequence: chr6:43769178..43769297 (hg38)
Cut-offs: matrix similarity = 0.97; core similarity = 1.00; Exclusion of low-quality matrices
Fig. A: MATCHTM in TRANSFAC® Release 2022.1; Profile: vertebrate_non_redundant.prf
Fig. B: MATCHTM public 1.0 with library of positional weight matrices of TRANSFAC® public from 2005; Profile: vertebrate matrices
More evidence towards the Professional version excellence
To be completely convinced that TRANSFAC® Professional is a must-have for your gene regulation studies, watch this video from geneXplain’s CEO Dr. Alexander Kel, where he demonstrates the difference in the analysis of COVID-19 associated SNPs list, when site search is ran on the matrix libraries of TRANSFAC®Public, TRANSFAC® Professional, and JASPAR databases:
Want to get TRANSFAC® Professional? Select the package of your interest and request its price here.
For any questions on our tools and databases, please contact us via info@genexplain.com


