Promoter analysis of RNA-seq preprocessed data in MATCH Suite
Promoter analysis of RNA-seq preprocessed data in MATCH Suite
Having done the pre-processing of your RNA-seq data, you can submit the obtained list of differentially expressed genes (DEGs) to the gene set analysis provided by the MATCH Suite tool. MATCH Suite scans the promoter regions of your DEGs and finds transcription factors that regulate your genes. It takes into account both: site enrichment and site combinatorial analysis and provides you with a reliable model of regulation of your target genes. MATCH Suite is the toolbox of TRANSFAC 2.0 that allows you to explore the functional enrichment of your gene set, find only transcription factors (TFs) expressed in the tissue of your interest, filter the predicted TFs by those intersecting with conservative regions of the genome and much more!
MATCH Suite accepts lists of 20 to 2000 genes coming in Ensembl ID, Entrez ID or Gene symbol format. Having uploaded your list of DEGs, you can proceed to specifying the studied tissue, the promoter range, and the GO terms of your interest. The tissue selection will allow to use tissue-specific promoters from the FANTOM5 database (where available) and identify the transcription factors regulating your gene set that are known to be expressed in the selected tissue. The GO terms selection will to optimise your input gene list and submit only genes belonging to the selected GO categories to further analysis (if this parameter is selected, an interactive tree map of GO biological processes, cellular components or molecular functions will be constructed for your input gene set depending on your selection; you can click on the categories of your interest and leave only genes belonging to them for the performed analysis).
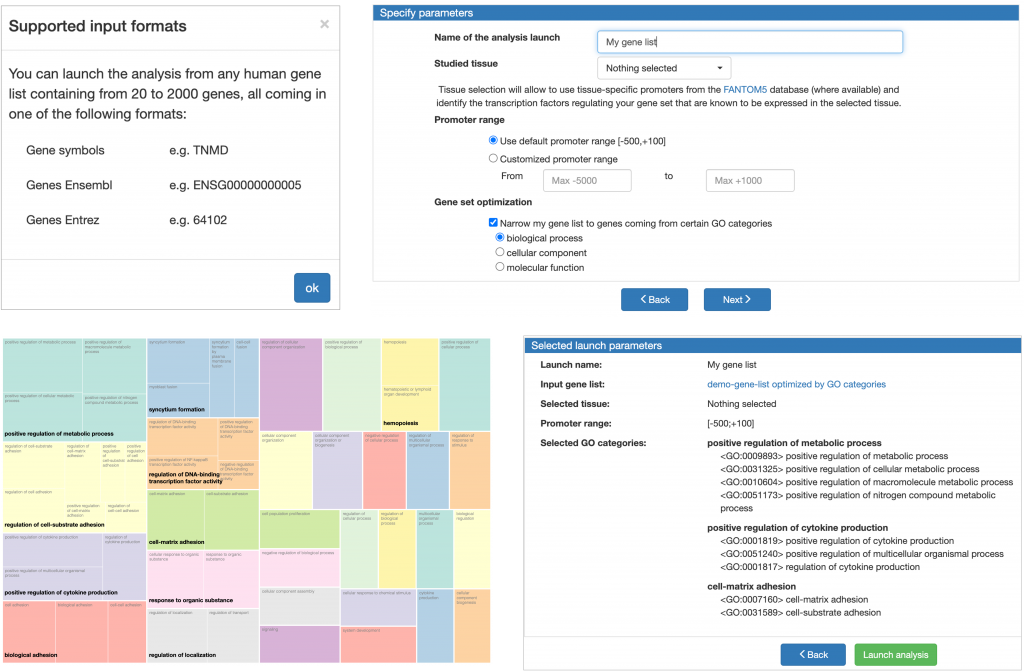
The analysis results of MATCH Suite can be viewed in the interactive interface or observed in the analysis report automatically generated for each analysis launch.
The interactive results visualisation section of the MATCH Suite provides you with dynamically filterable tables of the eventually identified TFs and respective matrices (binding models). The genes from the input set are compiled to a dynamically updating table, which demonstrates the regulation of these genes by respective TFs and binding motifs. Interactive genome browser visualisation of the identified sites in the promoters of your input genes is providing intuitive overview of the obtained results and various customisable filters applied.
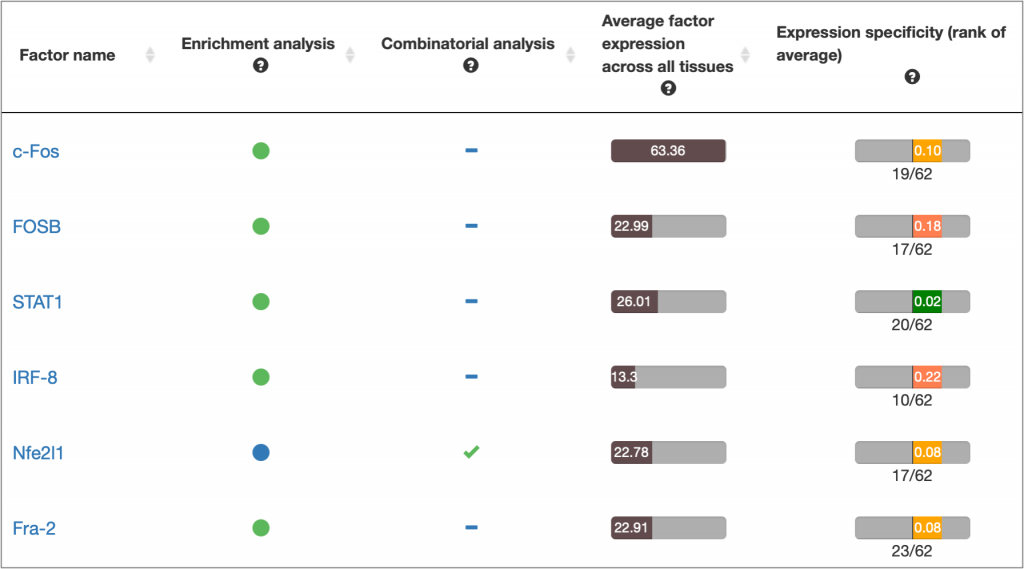
MATCH Suite gene set analysis result: Factor Overview table example:
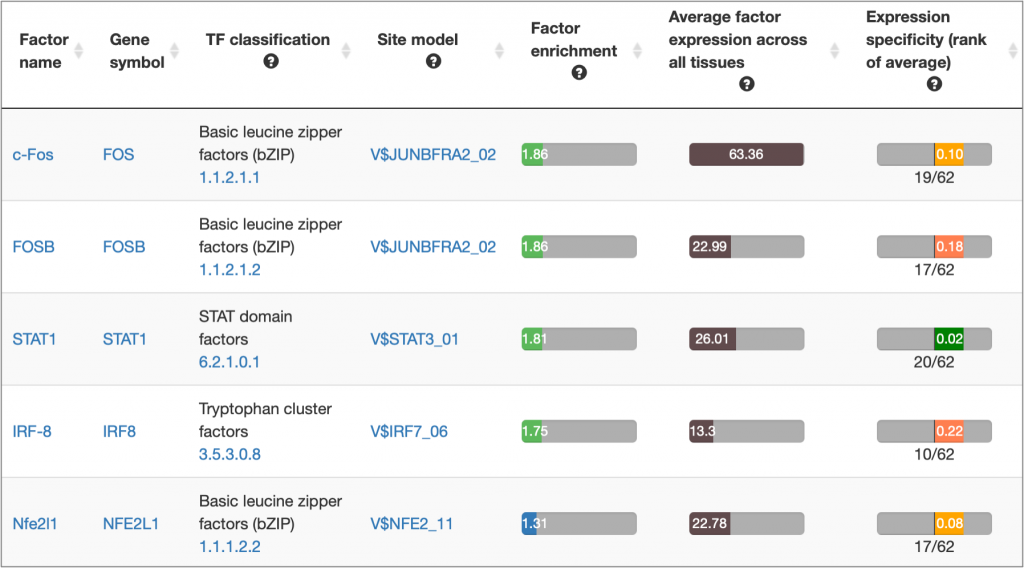
MATCH Suite gene set analysis result: Factor Pro table example:
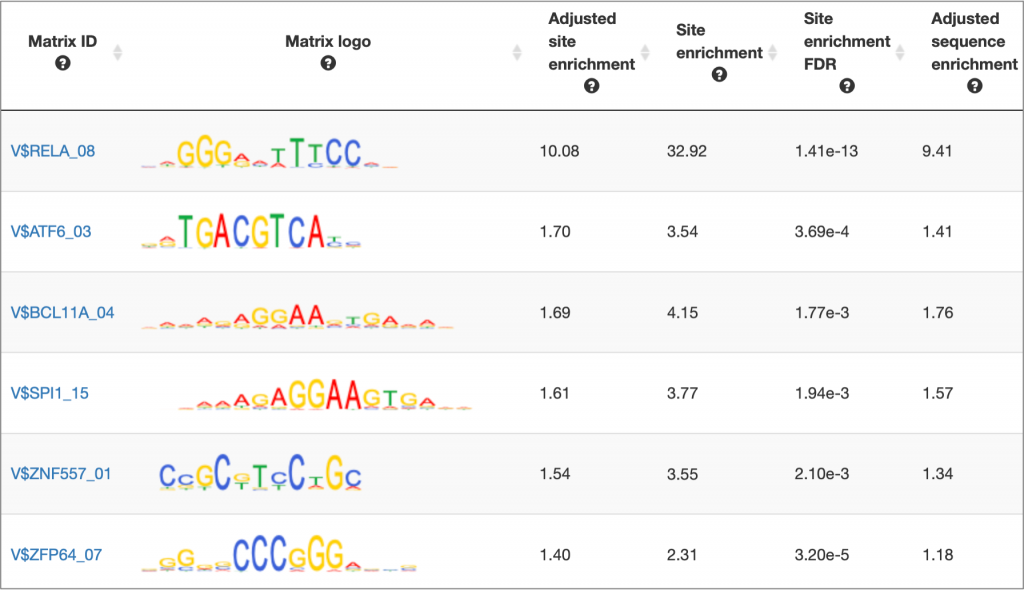
MATCH Suite gene set analysis result: Matrix table example:
MATCH Suite gene set analysis result: Genes table example:
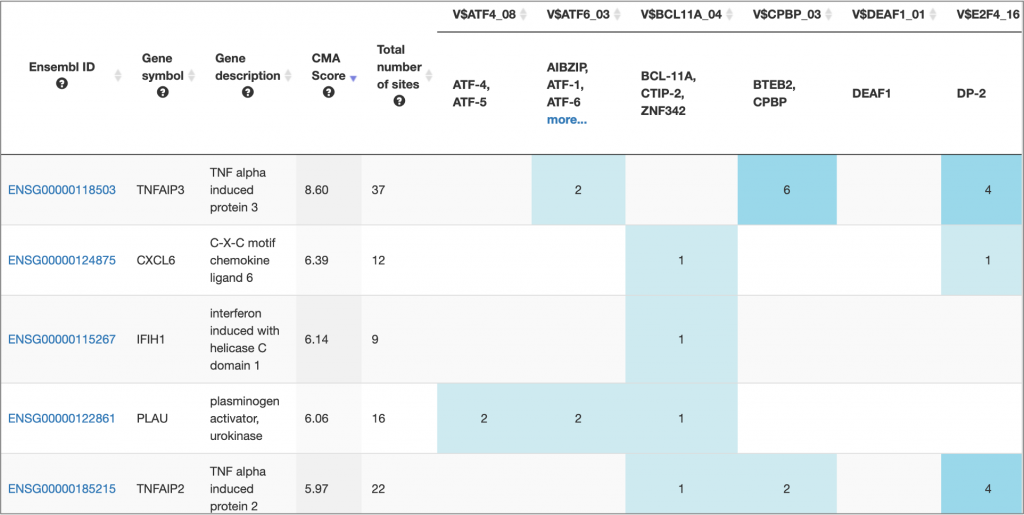
MATCH Suite gene set analysis result: Genome browser visualisation example:
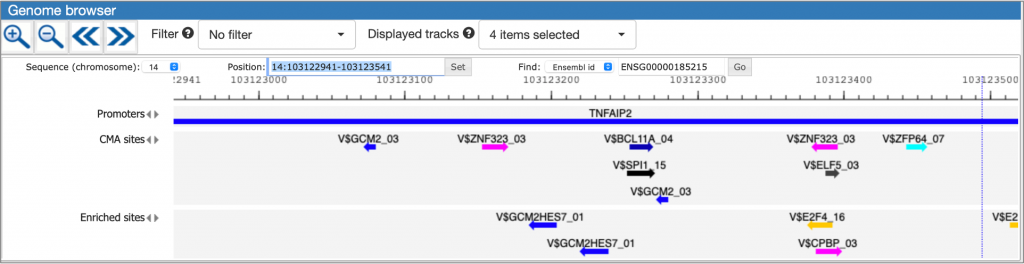
The interactive results visualisation interface of MATCH Suite allows dynamic filtering of the identified transcription factors and their binding models (matrices) by a number of parameters, including additional tissue selection, as well as cut-offs selection for Adjusted site enrichment and Adjusted sequence enrichment parameters, or filtering of the binding sites displayed in the Genome Browser, which allows to leave only sites intersecting with the conservative regions of the genome or those intersecting with the enhancer regions. You can view details on these operations in the MATCH Suite User Guide, pages 13-21).
The MATCH Suite also automatically generates a comprehensive report on rigorously filtered transcription factors regulating your input genes, you can explore an example report generated by the MATCH Suite below:
MATCH Suite can be launched from a direct product link at geneXplain databases portal (subject to TRANSFAC licensing).
More details about MATCH Suite can be found on the MATCH Suite product page.



