Structure
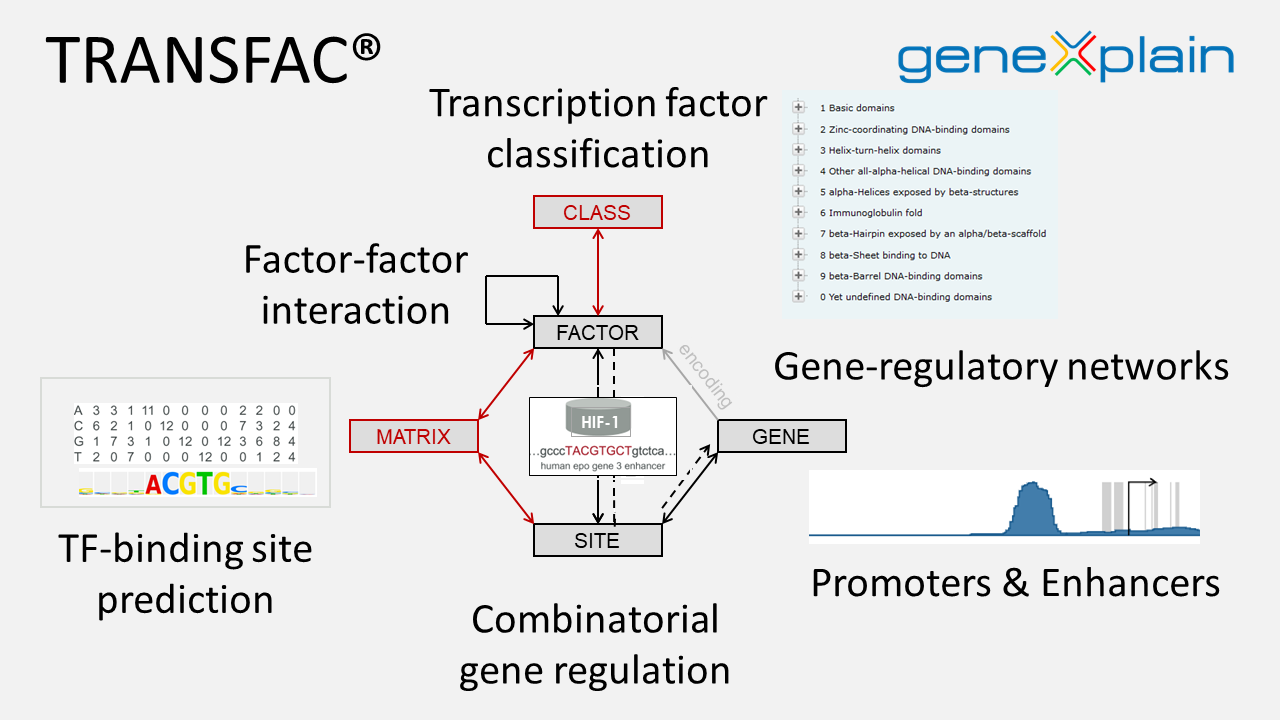
At the center of TRANSFAC® are transcription factors and their DNA binding sites based on experimental evidence extracted from scientific publications. The interlinked factor and site entries are captured in the respective flat files. Interactions between two or more factors are also included. TF-binding sites can be with or without a connection to a gene. Genomic sites are mapped to promoter sequences in the TRANSProTM module of TRANSFAC®. Besides linkage of factors via binding sites to regulated genes, monomeric factors are also linked to their encoding genes. In some cases this may also lead to autoregulatory loops, where a factor regulates the expression of its own gene. Based on gene-factor and (indirect) factor-gene links, gene-regulatory networks can be extracted.
