Structure
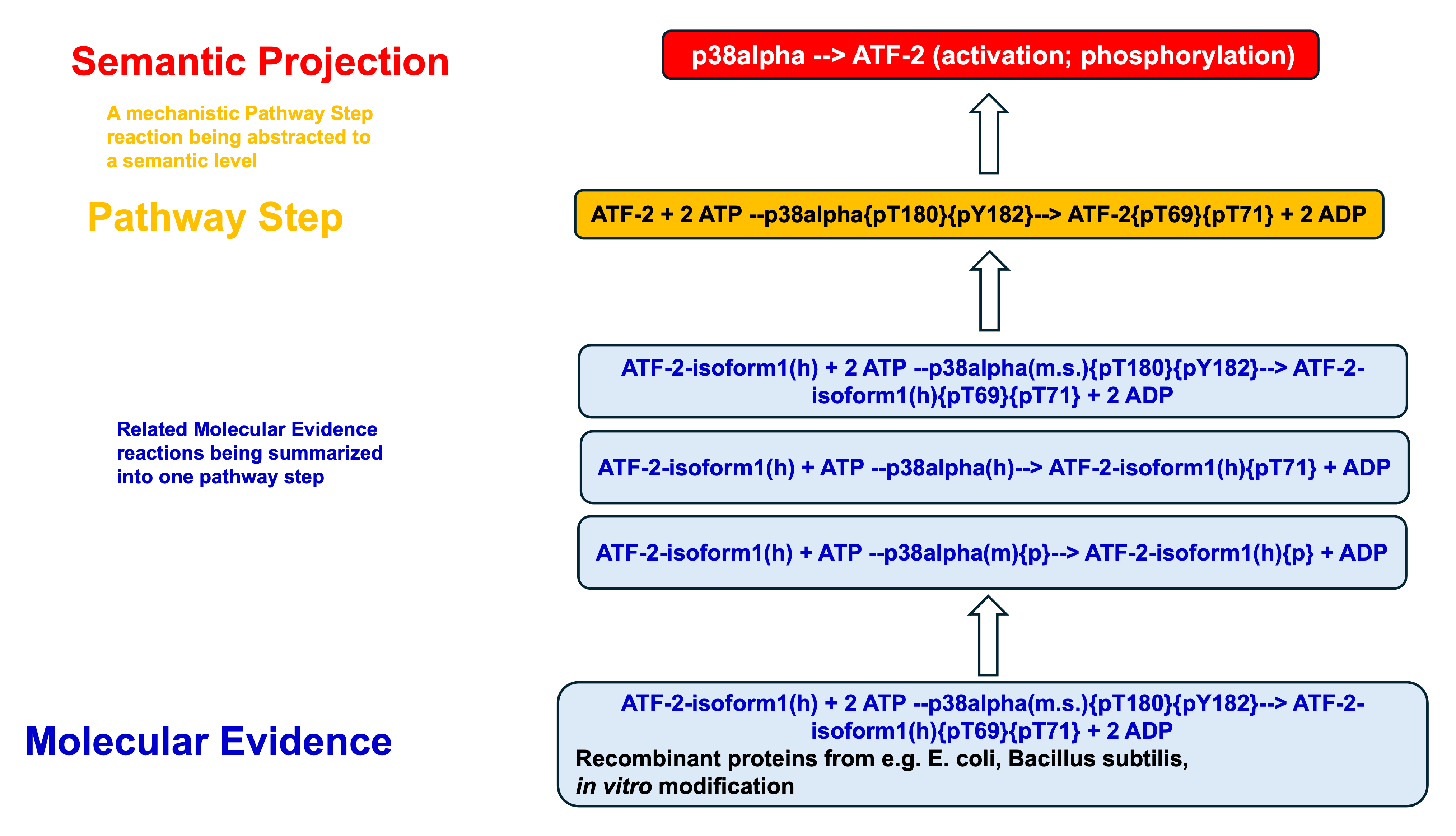
Reaction hierarchy in the TRANSPATH® database on molecular pathways.
TRANSPATH organizes the information about genes/molecules and reactions according to multiple hierarchies. Its sophisticated structure makes it one of the scientifically best conceptualized pathway resources, suitable for multi-purpose uses. It is complemented by one of the richest corpora of pathway data available among all public domain and commercial sources, all manually curated by experts.
Individual reactions are documented with all experimental details, in a strictly mechanistic way that includes all reaction partners and the taxonomic origin of each molecule as reported in the published experiment (“molecular evidence level”). All evidences for a certain pathway step are accumulated to provide a more comprehensive and complete picture (“pathway step level”). On top, a semantic view is provided, which focuses on the key components only and omits mechanistic details as well as small abundant molecules (“semantic projection”). Complete networks and pathways are built from molecules and their reactions.
To consider the heterogeneity of information given in the original publications, TRANSPATH transparently but precisely differentiates protein molecules according to:
-
- their relatedness within one genome
Information can be specifically retrieved regarding:
- their relatedness within one genome
(a) specific individual proteins,
(b) all products of a certain gene (isoforms),
(c) different family relation levels (e.g., paralogs);
-
- their relatedness between different genomes (orthology)
- their association and modification status
(a) protein complexes are specified with their exact composition;
(b) post-translational modifications are given with their exact positions in the protein.
