Step 1: Promoter analysis
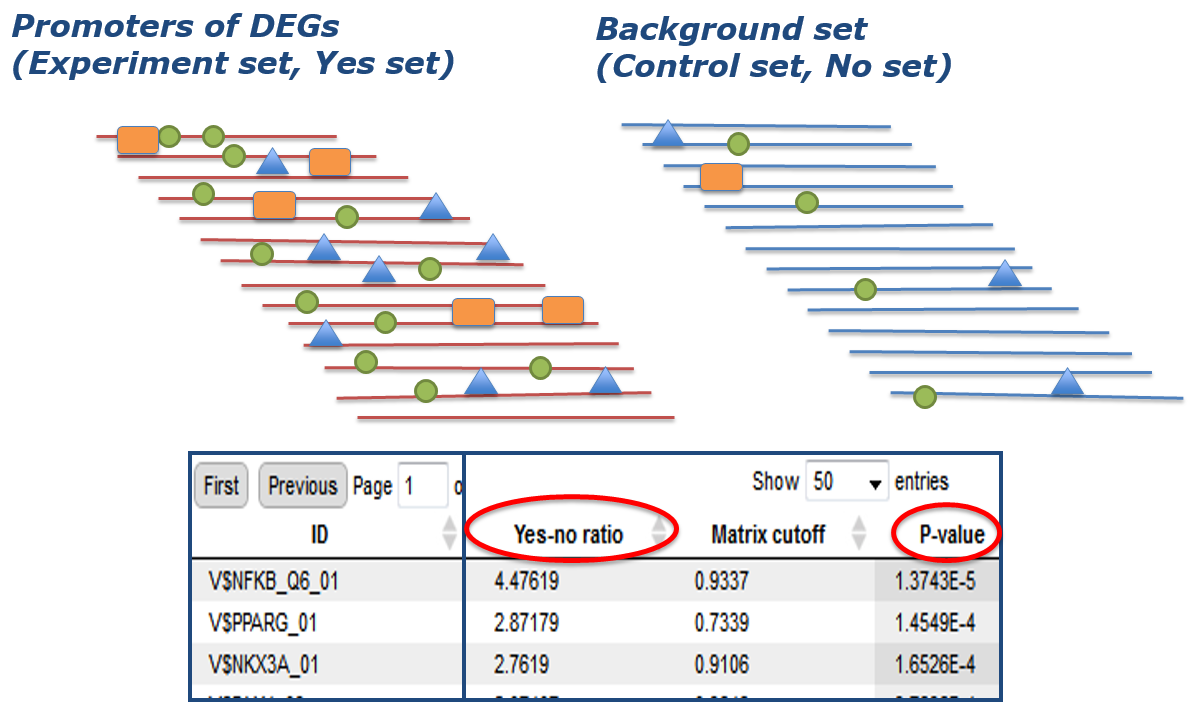
First, potential transcription factor binding sites (TFBSs) are identified in all promoters of the DEGs of your experiment (Yes set) as well as in a negative control set (No set). This is the usually done with a library of position-specific scoring or positional weight matrices (PSSMs or PWMs).
We recommend to apply the most comprehensive matrix library available, the TRANSFAC® database, and using the MATCHTM algorithm for the sequence analysis.
Next, out of all these potential transcription factor binding sites (TFBSs), those that are characteristic for the DEG set under study are identified. This is done by rigorously determining their enrichment in the Yes- compared to the No set.
Next, out of all these potential transcription factor binding sites (TFBSs), those that are characteristic for the DEG set under study are identified. This is done by rigorously determining their enrichment in the Yes- compared to the No set.