Advanced promoter analysis
Therefore, the geneXplain platform provides an empirical way to identify the specific combination of sites that characterizes a given set of co-regulating promoters.

Complex promoter analysis, visualization of transcription factor binding sites (TFBSs) constituting a “promoter model”. (Click Image for an enlarged view.)
These specific combinations, also called “promoter models”, can be further used for screening genomic sequences or promoter databases. A comprehensive collection of mammalian promoters comes along with the TRANSFAC® database in its TRANSPRO section. The density of model matches is visualized by graded shading.
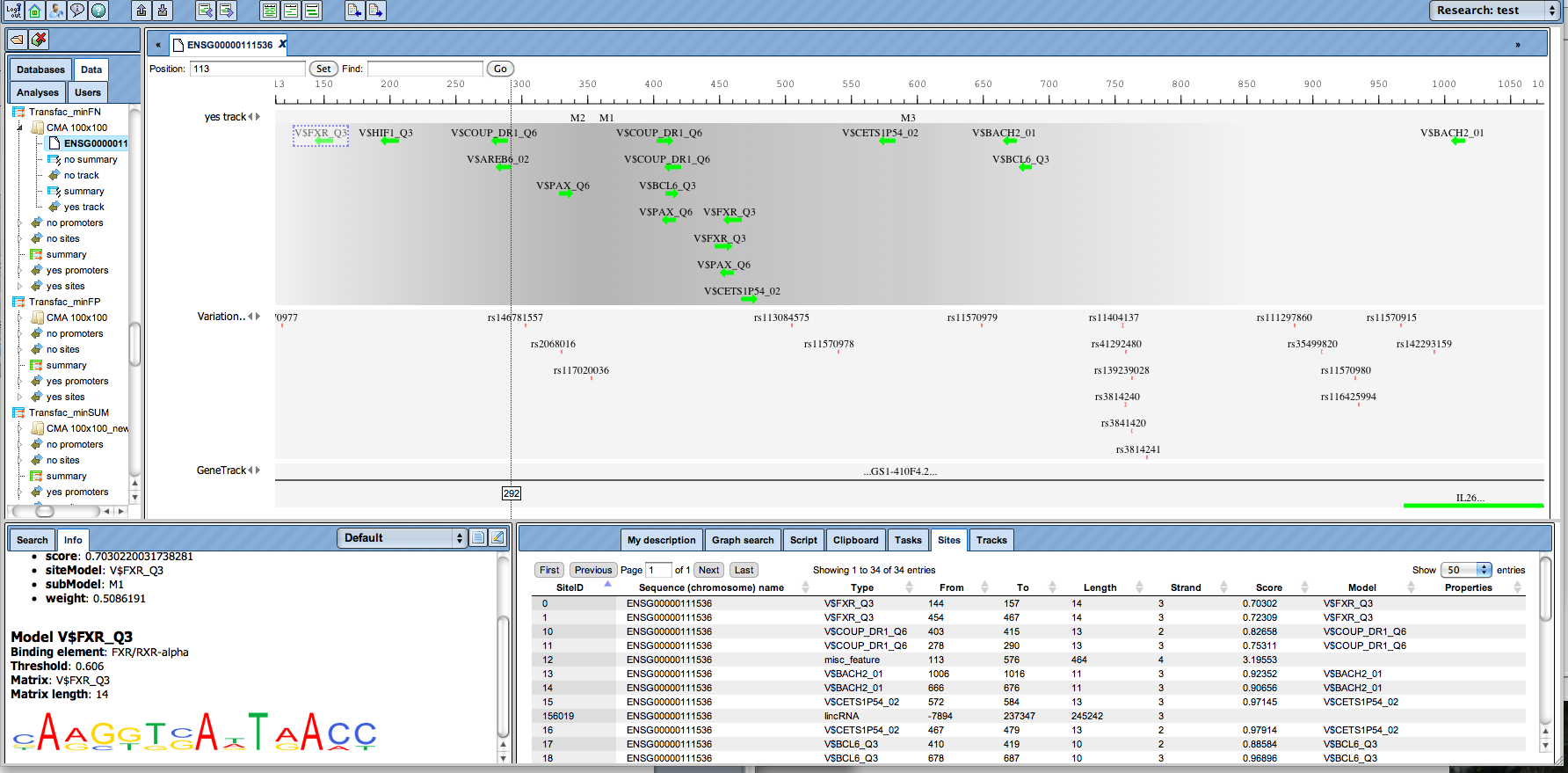
Promoter analysis for matches with a model comprising a set of transcription factor binding sites (TFBSs). (Click Image for an enlarged view.)
