MyPathSem
A data integration platform for generating patient-specific signaling pathways for personalized treatment decisions in clinical applications
Eine Datenintegrationsplattform für die Generierung von Patienten-spezifischen Signalwegen für individualisierte Behandlungsentscheidungen in klinischen Anwendungen


A collaborative project funded by the German Federal Ministry of Education and Research (BMBF) in the funding program
“i:DSem – Integrative Datensemantik in der Systemmedizin”
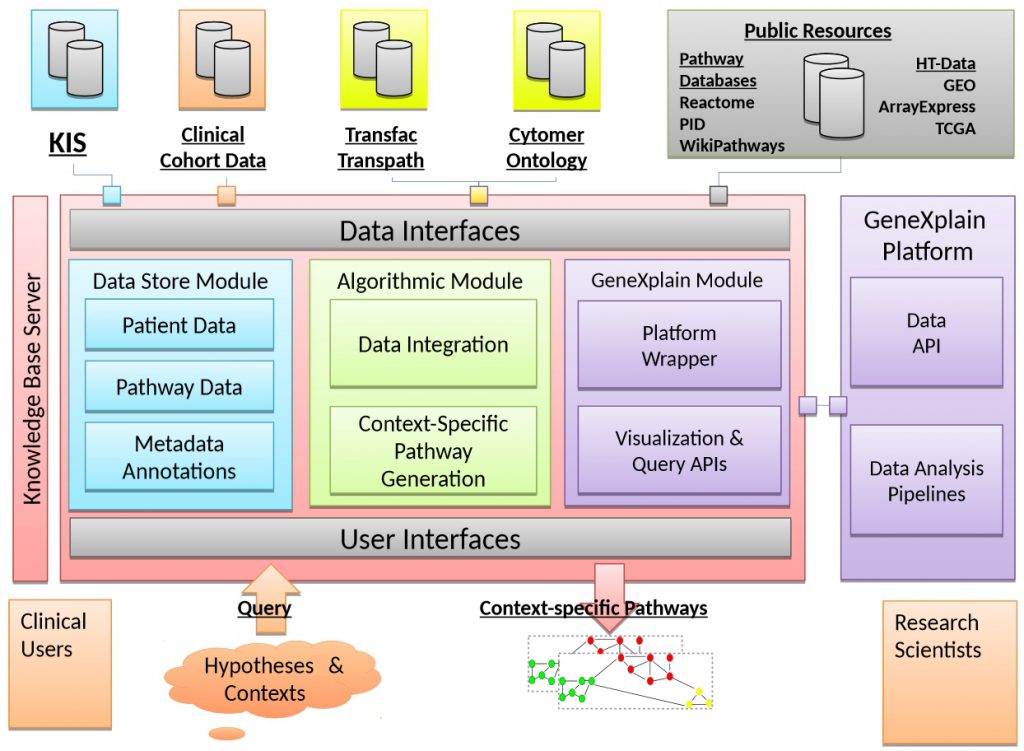
- to link clinical information systems with patient-specific Omics data;
- to generate a tool for data integration and easy access in a clinical environment;
- to collect information from public pathway and literature databases;
- to develop methods to generate context-specific pathways from individual patient data;
- to apply the new tools on data from colorectal and metastatic cancer;
- to test the utility in a clinical research setting; to integrate the tools into the geneXplain platform to make it available to a broader community; and, thus, to reduce the gap between patient centered routine documentation and ontology-driven pathway and gene annotation and establish a seamless data-flow from single patient data to Systems Medicine.
It is geneXplain’s task to contribute a user-friendly computer platform and to provide a prototypic version of an integrated toolbox that can be disseminated among research clinicians for systematic and large-scale validation of the project achievements. GeneXplain will also help disseminating the toolbox through its commercial channels, thereby contributing to the required sustainability of the project.