HumanPSD™+TRANSPATH®
Welcome to HumanPSD™+TRANSPATH®!

The Human Protein Survey Database (HumanPSD™) is a rich information resource connecting pathways with targets, drugs and clinical trials. It is greatly enhanced by the integrated TRANSPATH® database of mammalian biological pathways and networks.
The particular value of HumanPSD™+TRANSPATH® lies in the extensive documentation of protein molecules as drug targets and biomarkers. By connecting clinical phenotypes (diseases) through drugs with their targets, and further to the pathways they are involved in, HumanPSD™+TRANSPATH® supports you in making surprising discoveries.

Key Benefits:
- You can use HumanPSD™+TRANSPATH® as encyclopedia of signal transduction or metabolic pathways or for visualization of pathway connections between any molecules of interest.
- Quickly access detailed reports for individual genes, proteins, miRNAs, diseases, and drugs without time consuming literature search.
- Uncover biologically relevant connections between seemingly disparate genes, diseases, and drugs.
- Identify and rank potential therapeutic targets based on known functional characteristics.
- Explore canonical pathways and build custom protein networks, overlaying known disease and drug associations.
- Richest corpora of pathway data available among all public domain and commercial sources, all manually curated by experts.
- Enjoy the easy online access to HumanPSD™+TRANSPATH® or enjoy the privacy of a local installation on your server.
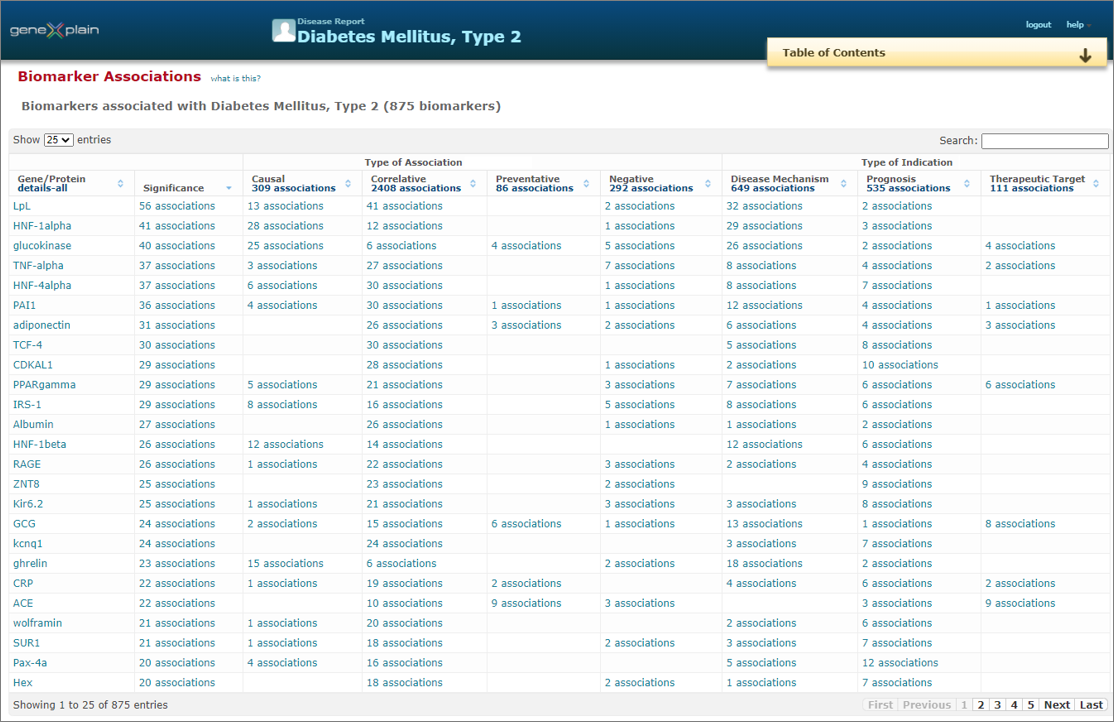
Disease / biomarker association
A tabular summary of literature-derived relationships between human genes and gene products with human diseases is given. These associations are clearly sorted according to their type, e.g. whether a gene/protein has a causal relationship with a disease to develop, or whether it is merely correlative, etc.
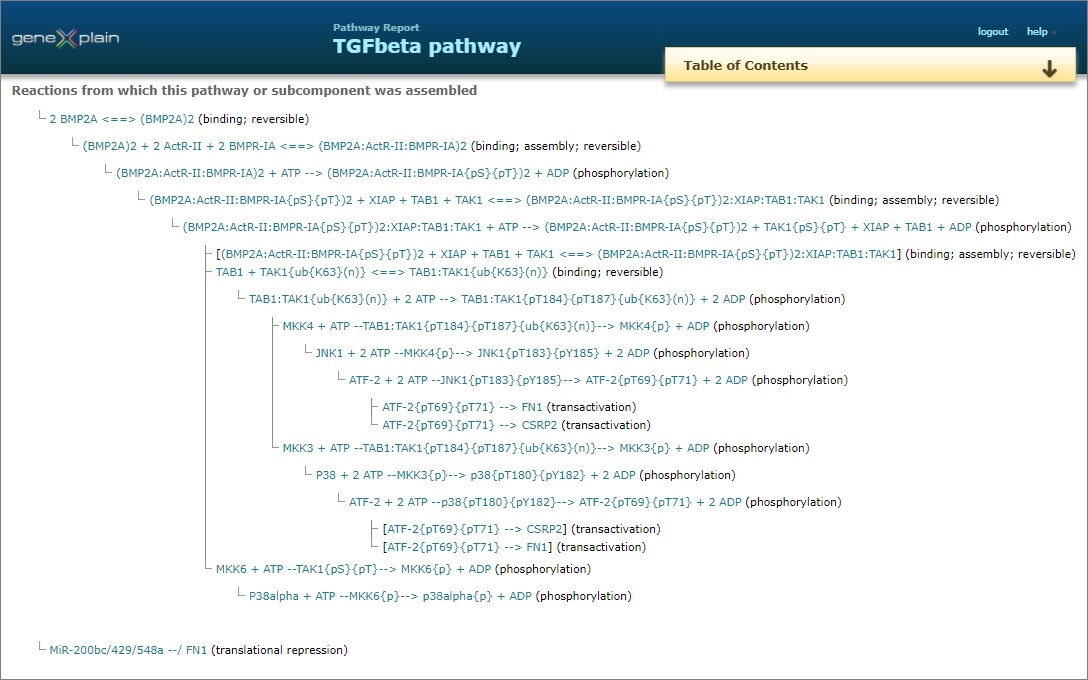
Networks & pathways
Complete networks and pathways are built from molecules and their reactions. To consider the heterogeneity of information given in the original publications, HumanPSD™+TRANSPATH® transparently but precisely differentiates protein molecules according to:
• their relatedness within one genome (isoforms/paralogs)
• their relatedness between different genomes (orthologs)
• their association and modification status (complexes, phosphorylations, etc.)
Disease similarity maps
The in-built disease similarity maps provide you with the unique info on affinity of various pathologies based on the genes that are involved in their acquisition and development. More info about disease-disease associations presented in HumanPSD™+TRANSPATH® can be found here.
