Genomic variants analysis in Genome Enhancer
Genomic variants analysis in Genome Enhancer
Genome Enhancer is a fully automatized pipeline for analysing patient omics data and studying the molecular mechanism underlying the observed pathology. Genome Enhancer identifies prospective drug targets and associated treatments from the input omics data, supported data formats include genomic variants data.
We provide a number of demo reports demonstrating how Genome Enhancer results would look like on various inputs. Full list of all available demo reports can be found here. Below you will find the demo reports that demonstrate the results of Genome Enhancer analysis ran on the genomic patient data.
Demo report ‘SNP associated with Diabetes Mellitus — Genomics, SNP list’ – demonstrating how a list of SNPs can be analyzed in Genome Enhancer:
SNP-associated-with-Diabetes-Mellitus-Genomics-SNP-list
Demo report ‘Colorectal Cancer (Personalized patient data) — Genomics, VCF’ – demonstrating how genomics data in VCF format can be analyzed in Genome Enhancer:
Colorectal-Cancer-Personalized-patient-data-Genomics-VCF
Demo report ‘MTB (Molecular Tumor Board) report example for colorectal cancer patient — Genomics, VCF’ – demonstrating how genomics data in VCF format can be interpreted by Genome Enhancer in terms of the direct variant-drug associations taken from the clinical interpretation of patient variants databases:
MTB-Report-Colorectal-Cancer-Personalized-patient-data-Genomics-VCF
Other Genome Enhancer demo reports, which provide more examples of genomics patient data analysis coming from different diseases, can be found here.
How does it work
In the first step of this analysis the transcription factors that regulate the mutated genes (genes carrying sequence variations in the analyzed pathology) are identified with the use of the TRANSFAC® database of transcription factors binding sites.
The second step searches for common master-regulators of the identified transcription factors by building a personalized signal transduction network of the studied pathology using the TRANSPATH® database of mammalian signal transduction and metabolic pathways.
The identified master regulators are prospective drug target candidates. They are used for further selection of chemical compounds that can bring therapeutic benefit for the studied clinical case. In this step the HumanPSD™ database is employed to identify drugs that have been tested in clinical trials. The cheminformatic tool PASS predicts small molecules that can affect the identified targets.
Finally, Genome Enhancer generates a comprehensive analysis report about the personalized drug targets identified for a certain patient, or a group of patients, and the drugs that may be effective in this case. You can view a number of Genome Enhancer demo reports here.
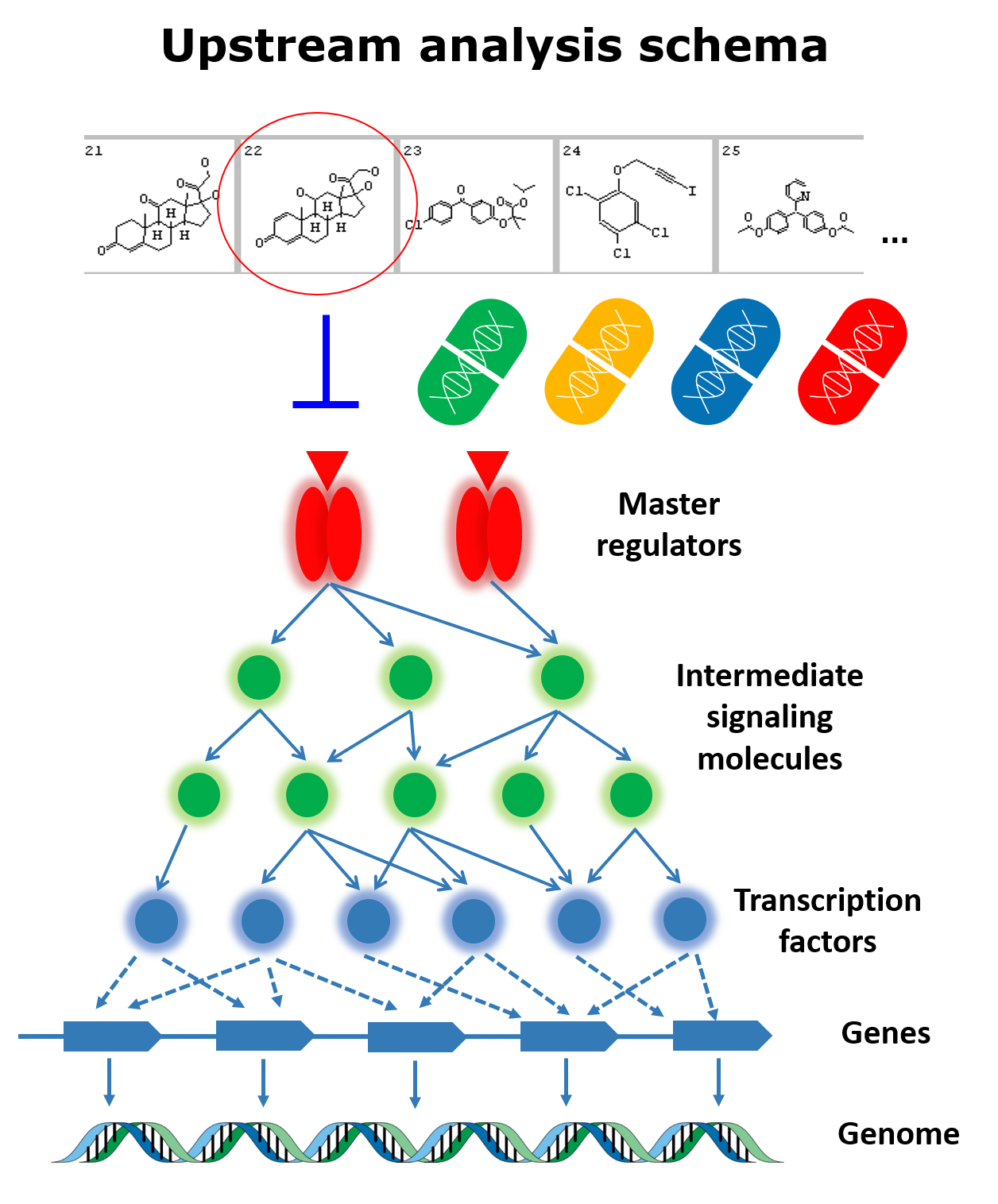
A detailed description of full Genome Enhancer analysis schema, including all steps of genomics data pre-processing, which are performed by the system for finding the mutated genes, can be found on this page. Genomics data can be loaded to Genome Enhancer in one of the following formats:
-
- Table data with SNP identifiers, rs*
- FASTQ files
- VCF files
You are welcome to purchase a paid subscription for Genome Enhancer by requesting it here.
For further info please explore the Genome Enhancer product page.
For any questions please feel free to contact us via info@genexplain.com


