Workflow management
Sequential launching of particular analysis modules can be saved as a graphically represented workflow. Modules are shown as purple rectangles, and outputs of each step serve as inputs into the next analysis step. A workflow that is specific for a given data set can be easily constructed by drag and drop of the required analysis modules. In addition, Java scripts and R scripts can be added directly within the platform, for more specific requirements of the analysis.
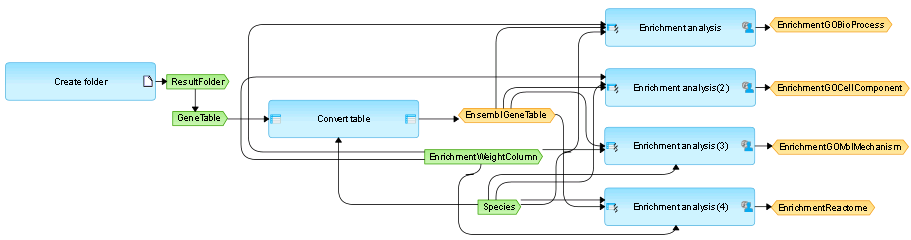
Workflow management in the geneXplain platform. (Click image to see the complete picture).
See demo workflows for a collection of executable workflows: no registration required!
