HumanPSD™ release 2024.1
The Human Proteome Survey Database (HumanPSDTM) with focus on human proteins as disease biomarkers and drug targets contains these new features:
- Biomarker and drug data update
The number of disease annotations increased to 408,644 and the number of unique gene/biomarker – disease assignments to 135,700.
Newly FDA-approved drugs (Sep 2023 – Mar 2024), their indication, and their protein target information were added.
HumanPSD is available for online access fully integrated with TRANSPATH
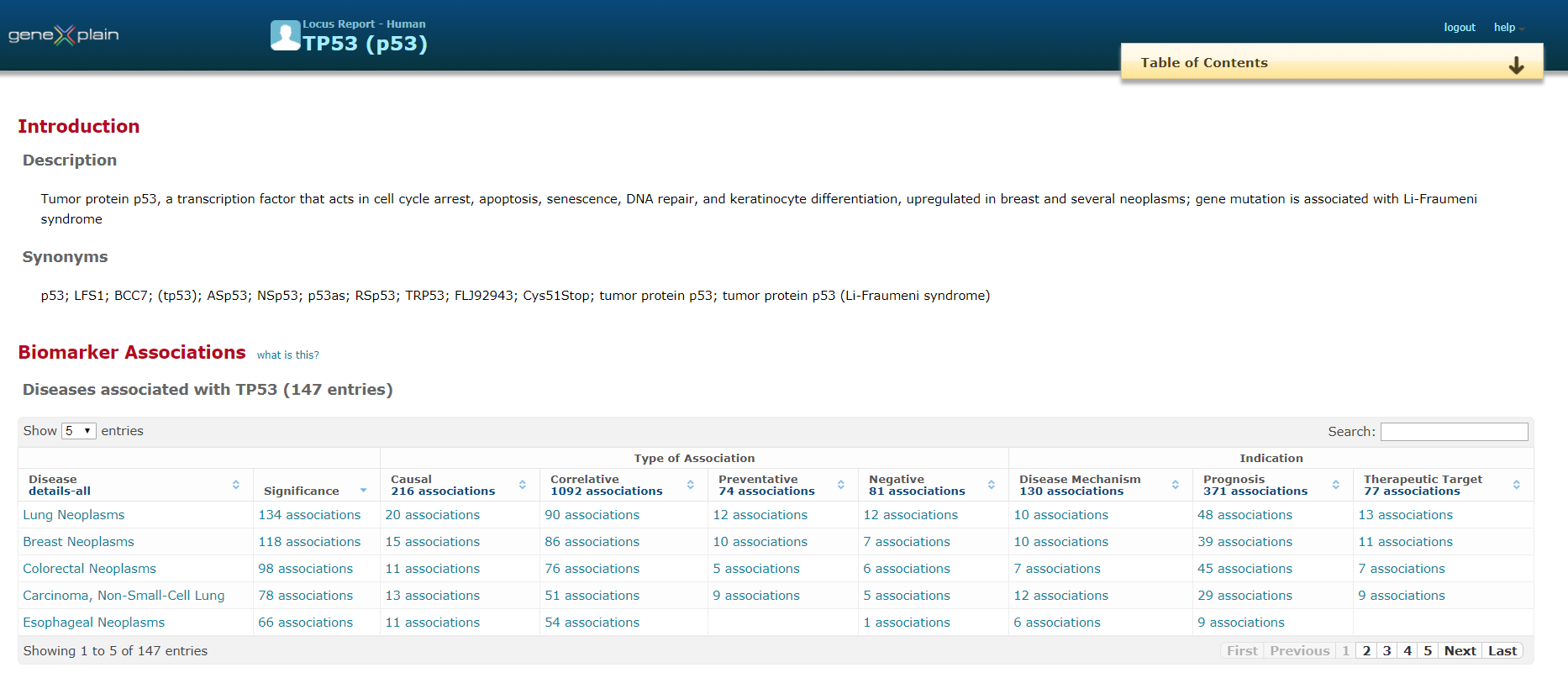
The basic information unit is a “locus report”, which summarizes the existing knowledge about the product(s) of a gene. It is part of a hierarchy, with individual proteins (isoforms such as splice variants) encoded by a gene at a level under the locus report, and summarizing features of the orthologs of human, mouse and rat origin at a higher level.
View demo report for IRF1 gene
HumanPSDTM Statistics (download)
HumanPSDTM Features (download)
HumanPSD FlyerTM (download)
Stegmaier, P., Krull, M., Voss, N., Kel, A.E., Wingender, E. (2010) Molecular mechanistic associations of human diseases. BMC Syst Biol. 4, 124. doi: 10.1186/1752-0509-4-124. PubMed.
Michael, H., Hogan, J., Kel, A., Kel-Margoulis, O., Schacherer, F., Voss, N., Wingender, E. (2008) Building a knowledge base for systems pathology. Brief. Bioinform. 9, 518-531. doi: 10.1093/bib/bbn038. PubMed.
Wingender, E., Crass, T., Hogan, J.D., Kel, A.E., Kel-Margoulis, O.V., Potapov, A.P. (2007) Integrative content-driven concepts for bioinformatics “beyond the cell”. J Biosci. 32, 169-180. PubMed.
Hodges PE, Carrico, P.M., Hogan, J.D., O’Neill, K.E., Owen, J.J., Mangan, M., Davis, B.P., Brooks, J.E., Garrels, J.I. (2002) Annotating the human proteome: the Human Proteome Survey Database (HumanPSD) and an in-depth target database for G protein-coupled receptors (GPCR-PD) from Incyte Genomics. Nucleic Acids Res. 30:137-141. PubMed.
HumanPSD is a trademark of QIAGEN GmbH.