What are Transcription factors?
Transcription factors (TFs) are the main regulators of gene activities. They play a central role in nearly every cellular process, thereby controlling practically any physiological phenomenon. Among them are the differentiation of pluripotent stem cells to highly specialized tissue cells, cellular responses to hormones, and sex development, just to name a few. Thus, TEs also play a key role in the etiology of
many diseases, for instance in all kinds of cancer.
About transcription
Let’s quickly review the transcription process before delving into the details of transcription factors. Transcription is the process of creating RNA from DNA and involves the use of enzymes called RNA polymerase and transcription factors. Transcription factors help the RNA polymerase bind to the promoter region of the gene and initiate the transcription process. In eukaryotes, the different types of RNA polymerases (I, II and III; two more in plant cells) are responsible for producing different types of RNA (1).
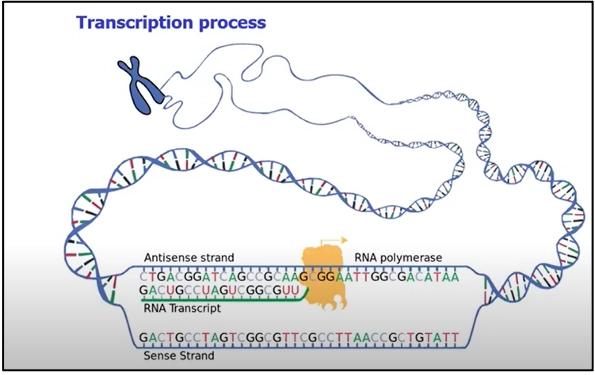
All three RNA polymerases require additional factors for specific promoter recognition (2). General transcription factors like TFIIA, TFIIB, TFIID, TFIIE, TFIIF, etc combine with RNA-polymerase II and the TATA box to initiate transcription at the transcription start site (TSS). Since many of them are multisubunit proteins, the pre-initiation transcription complex is a group of altogether 45 individual polypeptides assembled.
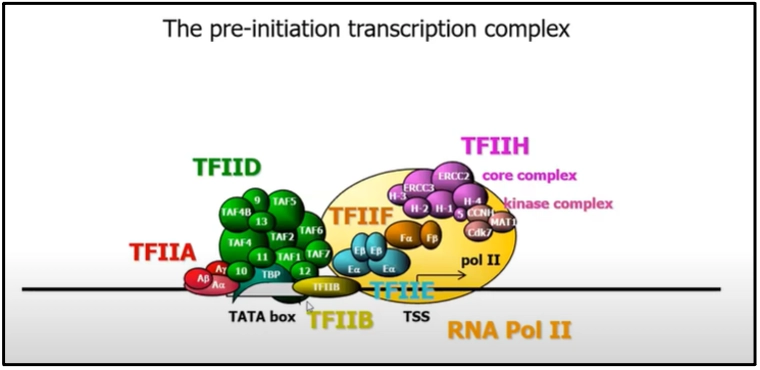
How do transcription factors operate?
In eukaryotic cells, transcription factors are nuclear proteins that associate with regulatory genome regions (promoters, enhancers and silencers), in most cases by binding to short DNA sequences of 5-25 base pairs (2).
TFs direct the RNA polymerase to the correct start site of transcription in a cell type and cell stage-specific way, frequently responding to extracellular stimuli. Most TFs bind to DNA in a somewhat relaxed sequence-specific way, thereby encoding the regulatory signals of the genome.
The function of TFs is to regulate − turn on or off − genes in order to make sure that they are expressed in the desired cells at the right time and in the right amount throughout the life of the cell and the organism (3).
Transcription factors may serve as disease biomarkers. According to the HumanPSD database (a manually curated database about diseases, biomarkers, pathways and drugs; release 2022.2 https://genexplain.com/humanpsd), transcription factors are proven to play an important role in close to 1600 diseases (4):
General structure of transcription factors
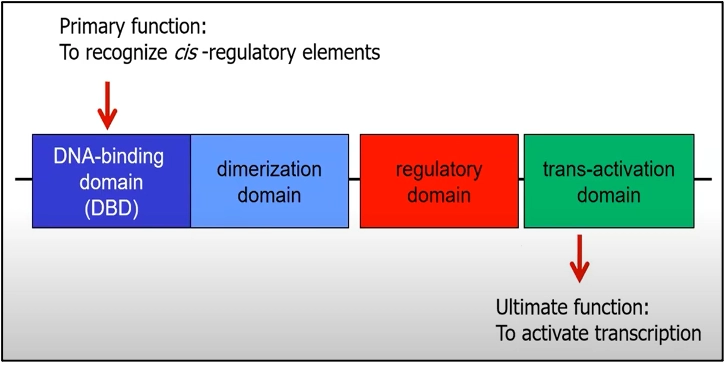
Most transcription factors bind directly to DNA. If we look at the structure of a typical transcription factor, we recognize 4 major domains:
- The DNA-binding domain: The main function of this domain is to recognize the cis-regulatory elements of the promoter.
- Dimerization domain: Many TFs form homo or hetero dimers and the kind of these dimers regulate the DNA-binding specificity.
- Regulatory domain: Some TFs need a regulatory domain to become active.
- Trans-activation domain (TAD): This domain is responsible to activate transcription, it contains binding sites for other proteins such as transcription co-regulators or to components of the transcription initiation complex, which helps in transcription activation. In general, for most factors, these domains are acidic domains, glutamine-rich domains, proline-rich domains, or isoleucine-rich domains.
Classification of transcription factors:
Transcription factors can be classified based on their DBDs. This is helpful in many ways:
- to establish a classification system for this important group of proteins
- to find a clue to decipher the protein DNA-recognition code
- to elucidate the evolutionary history of TFs
- to derive computational models for these DBDs that help identify orthologous TFs encoded by newly sequenced genomes and, thus, establish TF catalogs for these species.
Previous studies by Suzuki and Yagi (5) have proven that the protein-DNA recognition code is class specific and hence a comprehensive classification is required.
E. Wingender et al. (6, 7, 8) have done immense work to classify transcription factors. Inspired by the enzyme catalog a 4-level taxonomy was proposed (6) for TF classification. It comprises the ranks of superclass, class, family, and sub-family of the DNA binding domains.
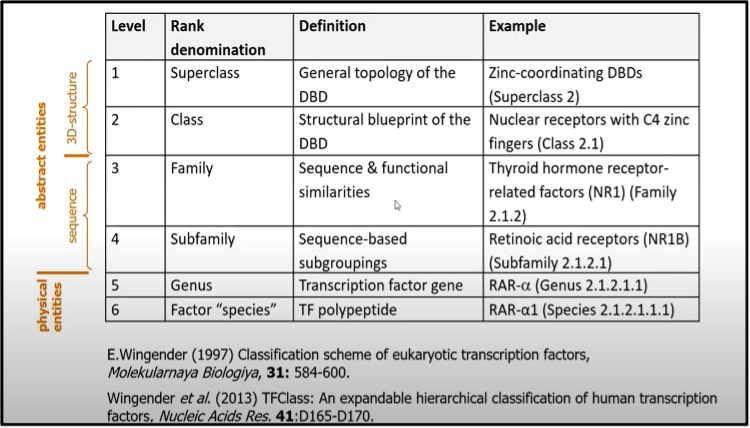
Superclasses are defined according to the general topology of the DBD and the principal mode of interacting with DNA. The general blueprint of the DBD i.e it’s folding and the way how it establishes the DNA interacting interface defines the class (9). So far nine super classes have been identified, comprising 40 classes and 111 families. Counted by genes, 1558 human TFs have been classified so far, or >2900 different TFs when including their isoforms generated by alternative splicing or protein processing events (8).
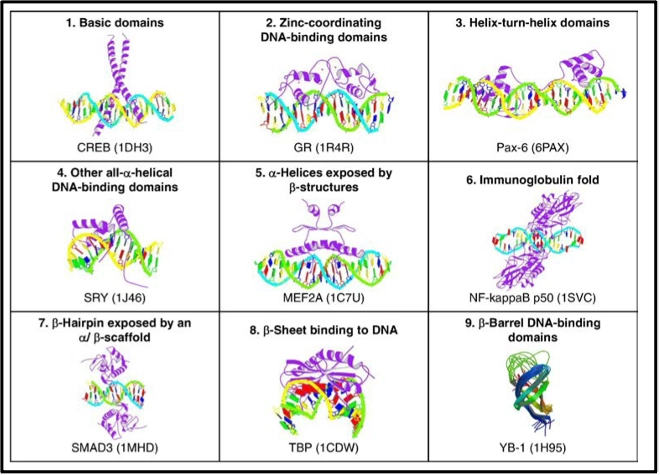
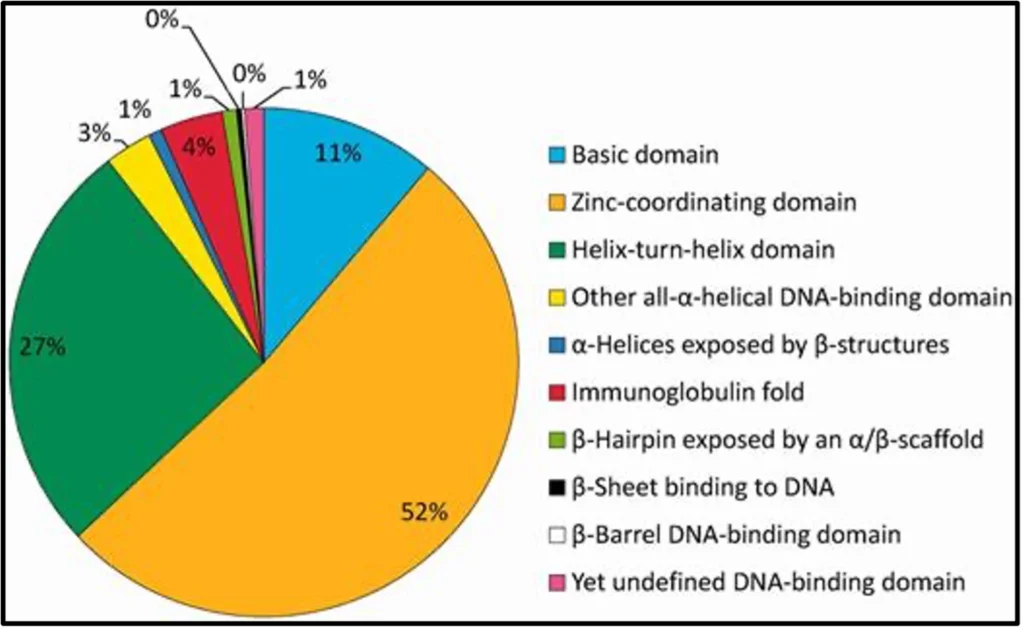
Among the nine super classes of defined DBDs, by far the largest is Superclass 2 (zinc-coordinating DBDs) to which 53% of all TF genes belong to, followed by helix-turn-helix (26%) and basic domain factor genes (11%). These three super classes have in common that an α-helix is exposed in such a way that it binds into the major groove of the DNA.
The complete link to the classification at all levels of all human transcription factors with free access to TRANSFAC factor entries can be found here.
The TRANSFAC database
As standard resource for (eukaryotic) transcription factors and their genomic interactions, the TRANSFAC database provides the relevant information (10). Besides of its encyclopedic value, its comprehensive library of positional weights matrices (PWMs) is used by a bunch of computational tools to predict potential TF binding sites in genomes and to put them into the proper biological context. The most advanced software in this regard is the MATCH Suite (11).
10 important points to remember about transcription factors:
- Transcription factors are proteins that bind to DNA regulatory sequences and modulate the rate of transcription.
- Most transcription factors are transcriptional activators, however some are also transcriptional repressors. Some may act either way, depending on the context of other TEs in the respective target genes.
- Many transcription factors function as master regulators and selector genes exerting control over many pathways and play a very important role in human diseases.
- The function of TFs is to regulate turn on and off genes. In some cases, TEs also form homo- or heterodimers by physically interacting with each other. This allows cross-talk between different signal transduction pathways at the level of gene expression.
- Four main domains can be identified in most TFs: DNA-binding domain
(DBD), a dimerization domain, a regulatory domain, and a transactivation domain. - The DNA-binding domain gives transcription factors the ability to bind to specific DNA-sequence elements, assemblies of which constitute enhancer or promoter regions.
- As per the latest TRANSFAC release (2022.2) there are a total of 48036 factors across all species, while there are close to 4000 human transcription factors encoded by nearly 2000 TF genes.
- Transcription factors can be classified into four general levels (superclass, class, family, subfamily) and two levels of instantiation (genus and molecular species).
- Transcription factor activation is a complex phenomenon and involves multiple pathways. Sometimes transcription factors may be activated by ligands such as glucocorticoids and vitamins A and D.
- As per HumanPSD release 2022.1, 1073 TFs belonging to different classes of DNA-binding domains are documented as biomarkers for 246 various cancer types, including neoplasms, carcinomas, sarcomas, lymphomas, leukemia, myeloma, granuloma, blastoma, glioma, and some others
References:
- Carter, R., Drouin, G. (2009) Structural differentiation of the three eukaryotic RNA polymerases. Genomics 94, 388-396. doi: 10.1016/j.ygeno.2009.08.011
- Matsui, T., Segall, J., Weil, P.A., Roeder, R.G. (1980) Multiple factors required for accurate initiation of transcription by purified RNA polymerase II. J. Biol. Chem. 255, 11992-11996
- Adcock, I.M., Caramori, G. (2009) Transcription Factors, Editor(s): Peter J. Barnes, Jeffrey M. Drazen, Stephen I. Rennard, Neil C. Thomson, Asthma and COPD (Second Edition), Chapter 31, Academic Press, p. 373-380, ISBN 9780123740014
- https://genexplain.com/humanpsd
- Suzuki, M., Yagi, N. (1994) DNA recognition code of transcription factors in the helix-turn-helix, probe helix, hormone receptor, and zinc finger families. Proc. Natl. Acad. Sci. USA 91, 12357-12361. doi: 10.1073/pnas.91.26.12357
- Wingender, E. (1997) Classification scheme of eukaryotic transcription factors. Mol. Biol. Engl. Tr. 31, 498-512
- Wingender, E., Schoeps, T., Dönitz, J. (2013) TFClass: An expandable hierarchical classification of human transcription factors, Nucleic Acids Res. 41, D165-D170
- Wingender, E., Schoeps, T., Haubrock, M., Dönitz, J. (2015) TFClass: a classification of human transcription factors and their rodent orthologs, Nucleic Acids Res. 43, D97–D102
- Wingender, E. (2013) Criteria for an updated classification of human transcription factor DNA-binding domains. J. Bioinform. Comput. Biol. 11, 1340007
- https://genexplain.com/transfac
- https://genexplain.com/match-suite
Get free multi-omics reports to your Email
