STAT5A in hepatocellular carcinoma (HCC)
TFCLass 6.2.1.0.5
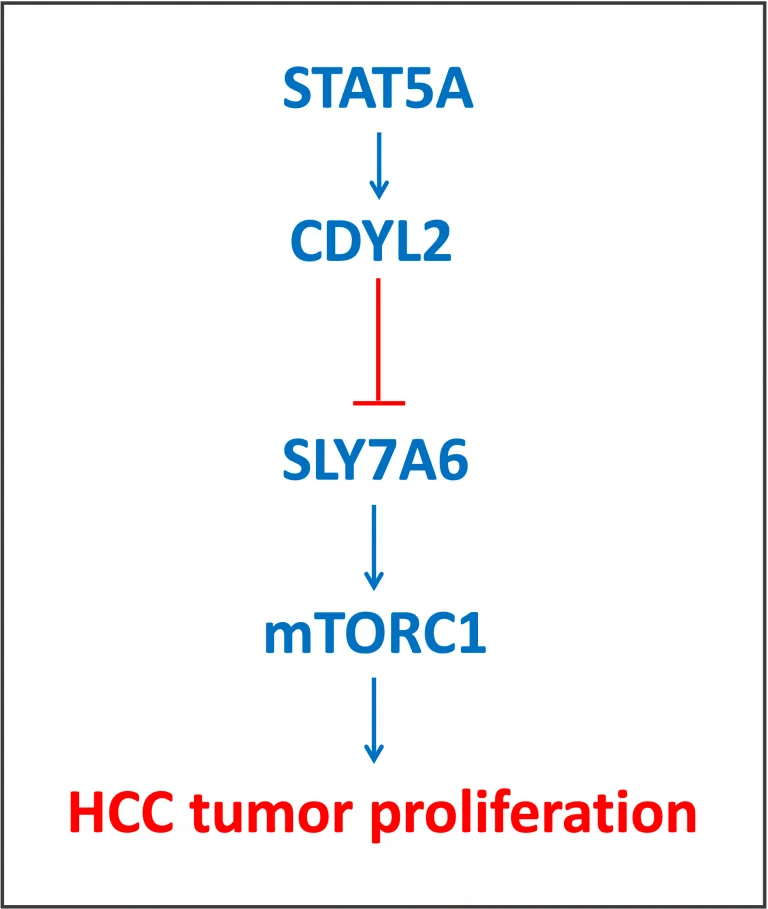
STAT5A was recently proven to up-regulate expression of CDYL2, a novel tumor suppressor in hepatocellular carcinoma (HCC; Chen, X. et al., 2022. Oncogene, 41: 2492-2504). The authors have shown that CDYL2 inhibits H3K4me3 histone methylation on the promoter of SLC7A6, an amino acid transporter. In its active status, SLC7A6 contributes to the activation of mTORC1, which is a well established regulator of cell proliferation in various tumors.
STAT5A was shown to directly bind its specific sites and activate expression of the CDYL2 gene in HCC cells. First, STAT5A binding sites were identified by the sequence analysis methods applying Jaspar, and then confirmed experimentally.
In summary, this study demonstrated activation of tumor suppressor CDYL2 by transcription factor STAT5A, and via SLC7A6 and mTORC1, this leads to inhibition of cell proliferation in HCC (Chen, X. et al., 2022. Oncogene, 41: 2492-2504).
STAT5A belongs to the class of STAT domain factors (TFClass link). STAT domain factors belong to a superclass of TFs known as “Immunoglobulin fold” (TFClass link).
STAT5A is known for its association with over 20 different diseases, the majority of them are various tumor types, as well as COVID-19 and HIV-infections. The top associated tumors are breast, prostatic, colonic neoplasms; several types of leukemia and lymphoma; glioblastoma and others (HumanPSD database). This study demonstrated the role of STAT5A in HCC (Chen, X. et al., 2022. Oncogene, 41: 2492-2504). A disease similarity map for hepatocellular carcinoma can be viewed here.
Expression of STAT5A protein varies significantly between the organs and tissues investigated. The highest expression level is observed in adipose tissue, bone marrow, lymph node, mammary gland, thymus and adrenal. Expression of STAT5A in normal liver is quite low, about 10-times lower as compared to its highest expression level in adipose tissue (HumanPSD database, Human Protein Atlas v20).
STAT5A is documented to bind its specific sites and activate transcription of a number of genes, including cyclin D1 (cell cycle regulator), CISH (cytokine-inducible signalling protein), DNMT3A (DNA-methyltransferase), transcription factor IRF1, tRNA synthetase WARS (TRANSFAC database). STAT5A was previously shown to repress the SLC7A11 gene (TRANSFAC database).
In TRANSFAC 2.0, there are 10 matrices for STAT5A. Among them, there are matrices based on the individual genomic binding sites, on the SELEX experiments, and on the ChIP-seq experiments. There are two family matrices for STAT factors. The core binding motif of STAT5A is TTCYYRGAAA, where Y can be T or C, and R can be A or G (TRANSFAC database).
STAT5A can be phosphorylated on particular serine or tyrosine positions especially in its C-terminal part (TRANSPATH database).
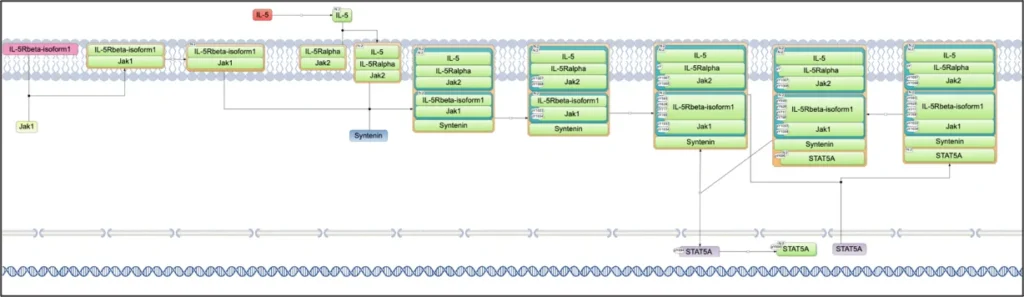
In the TRANSPATH database, there are several chains and pathways where STAT5A is involved, including Epopathway, ErbB4 mediated signalling, IL-3-, IL-5-, and PRL-pathways.
Here, the diagram of the IL-5 –> STAT5 chain is shown:
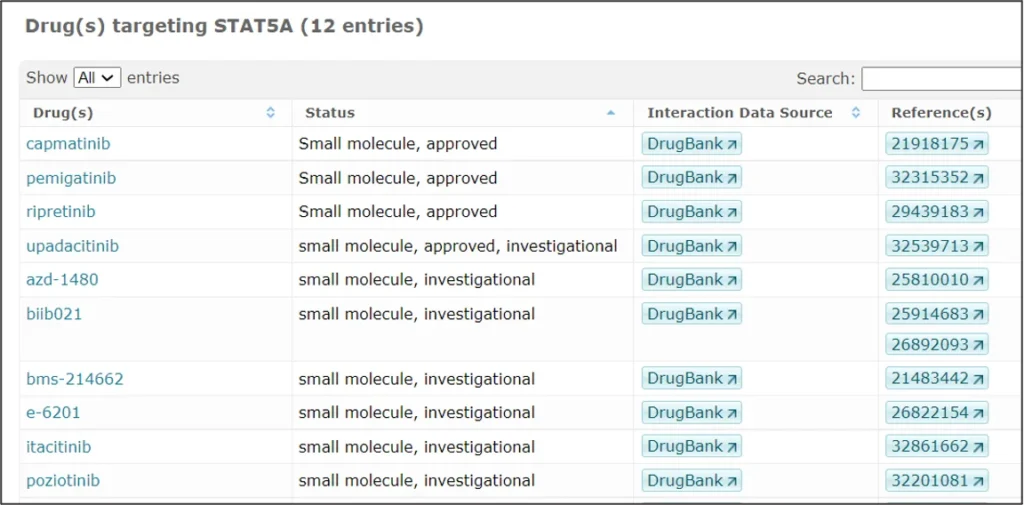
STAT5A is targeted by several drugs, out of those that are FDA approved and or under investigation in clinical trials, several examples are shown in the screenshot below (HumanPSD database):
Many more details can be found in the integrated database TRANSFAC + TRANSPATH + HumanPSD, here in the Locus Report for human STAT5A:
Open this report as a PDF file.
Chen X., et al., 2022. STAT5A modulates CDYL2/SLC7A6 pathway to inhibit the proliferation and invasion of hepatocellular carcinoma by targeting to mTORC1. Oncogene 41, 2492-2504. PMID: 35314791.
This page was published and last revised on 04.10.2022
Examples of some other transcription factors in cancer can be found here.
