RUNX3 and deltan-p63α in squamous cell carcinoma (SCC)
TFClass 6.4.1.0.3, TFClass 6.3.1.0.2.2
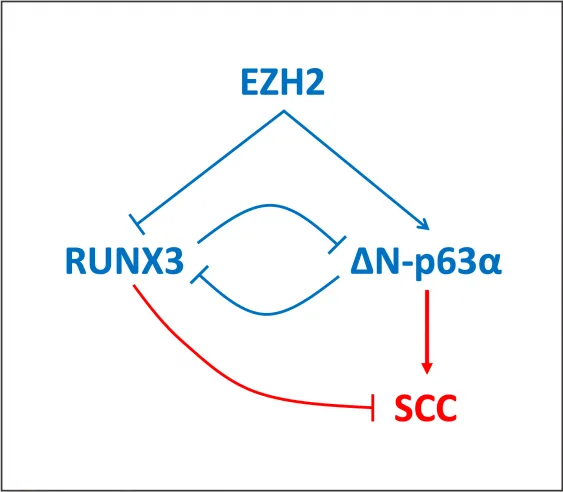
RUNX3 is a negative regulator of cell proliferation in several tumor types, and ΔN-p63α is an oncogenic isoform of TP63 transcription factor. ΔN-p63α isoform was previously shown to be highly expressed SCC. In a very recent publication, a repression of RUNX3 by ΔN-p63α was shown, which was activated by the oncogenic histone methyltransferases SETDB1 and EZH2 (Balinth, S. et al., 2022, Oncogene, v.41: 4130–4144). The authors have shown that tumor suppressor RUNX3, when active, significantly down-regulates expression of both SETDB1 and ΔN-p63α. Here, EZH2 was shown to repress RUNX3 and thus contribute to maintenance of a cancer stem cell phenotype in SCC. This study suggests that repression of EZH2 might be important for maintenance of active tumor suppressor RUNX3 in SCC (Balinth, S. et al., 2022, Oncogene, v.41: 4130–4144).
RUNX3, also known as PEBP2alphaC, CBF-alpha3, AML-2, together with RUNX1 and RUNX2, constitute the class of “Runt domain factors” (Wingender at al., 2018; TFClass link).
ΔN-p63α is one of 12 identified isoforms produced by the gene TP63. Among 12 isoforms, there are several DeltaN variants, they all lack N-terminal transactivation domain. All isoforms of TP63, together with 9 isoforms produced by TP53 and 9 isoforms produced by TP73, constitute the class of “p53 domain factors” (TFClass link). Both classes, Runt domain factors and p53 domain factors belong to a superclass of TFs known as “Immunoglobulin fold” (TFClass link).
RUNX3 is known for its association with over 60 different diseases, the majority of them are various tumor types, the top associated tumors are stomach, urinary bladder, pancreatic, esophageal, breast, lung neoplasms; hepatocellular carcinoma; esophageal squamous cell carcinoma and others (HumanPSD database). TP63 is associated with over 20 different tumor types, among them prostatic, lung, head und neck, urinary bladder, esophageal neoplasms; hepatocellular carcinoma; squamous cell carcinoma (HumanPSD database).
A disease similarity map for squamous cell carcinoma (SCC) is shown on the image aside and can be viewed in full-screen resolution here.
TP63 is expressed in only about one third of the tissues and organs investigated, with the highest expression level in skeletal muscle, skin, tongue and esophagus (HumanPSD database, Human Protein Atlas v20).
ΔN-p63α is documented to act as transcriptional repressor. Its DNA-binding domain is the same as that of p63-alpha, and therefore it can bind to the same sites. The absence of the transactivation domain results in gene repression. Site-specific DNA binding and repression by ΔN-p63α is known at least for several genes including CDKN1A (cyclin dependent kinase inhibitor 1A) and SFN (14-3-3sigma). In this publication, a repression of RUNX3 by ΔN-p63α was shown (Balinth, S. et al., 2022, Oncogene, v.41: 4130–4144).
Interestingly, ΔN-p63α is known to bind its specific site in the promoter of TP63 gene and repress its transcription; this creates a negative feedback loop: ΔN-p63α negatively regulates its own promoter. Through the same binding site TP63 gene transcription is positively regulated by TP53 providing an interplay between p53 domain factors. These facts and many more details can be found in the TRANSFAC database, in the Locus Report for this particular p63 isoform.
RUNX3 is a broadly expressed factor, it is identified practically in all the tissues and organs investigated, however its expression varies significantly, with the highest level identified in tonsil, bone marrow, spleen, lymph node, appendix, NK-cells, T-lymphocytes, thymus and small intestine (HumanPSD database, Human Protein Atlas v20).
In the TRANSFAC 2.0 database there are 15 positional weight matrices for RUNX3, either built on the individual genomic binding sites, based on a ChIP-seq dataset or SELEX experiments. Two of them are “family” matrices, where RUNX1, RUNX2 and RUNX3 are linked to. The binding motif is TGTGGT (RUNX3 Locus Report).
RUNX3 is acetylated by transcription co-activator CBP/p300 at several lysine residues within its DNA binding domain (TRANSPATH database). RUNX3 is a proven target molecule of at least two FDA-approved drugs, irinotecan and leucovorin (HumanPSD database, DGIdb).
Many more details can be found in the integrated database TRANSFAC + TRANSPATH + HumanPSD, for instance in the Locus Report for human RUNX3:
Open this report as a PDF file.
Balinth, S. et al., 2022. EZH2 regulates a SETDB1/ΔNp63α axis via RUNX3 to drive a cancer stem cell phenotype in squamous cell carcinoma. Oncogene, 41, 4130–4144. PMID: 35864175
Wingender E. et al., 2018. TFClass: expanding the classification of human transcription factors to their mammalian orthologs. Nucleic Acids Res. 46, D343-D347. PMID: 29087517
This page was published and last revised on 02.09.2022
Examples of some other transcription factors in cancer can be found here.
