NRF2/MAFG heterodimer in hepatocellular carcinoma (HCC)
TFClass 1.1.1.2.3, 1.1.3.2.2
Transcription factor heterodimer NRF2/MAFG has been identified to bind to its specific site and regulate transcription of aldo-keto reductase family 1 member C3 gene (AKR1C3) (Pan D. et al., 2022. Oncogene 41, 3846–3858). AKR1C3 is well known to contribute to a number of different cancers, and its association with hepatocellular carcinoma (HCC) is established, although the exact mechanisms were not elucidated yet. In this very recent publication, the authors have experimentally confirmed activation of AKR1C3 gene transcription by NRF2/MAFG heterodimers. The elevated expression of AKR1C3 leads to HCC progression, even independent of its enzymatic activity (Pan D. et al., 2022. Oncogene 41, 3846–3858).
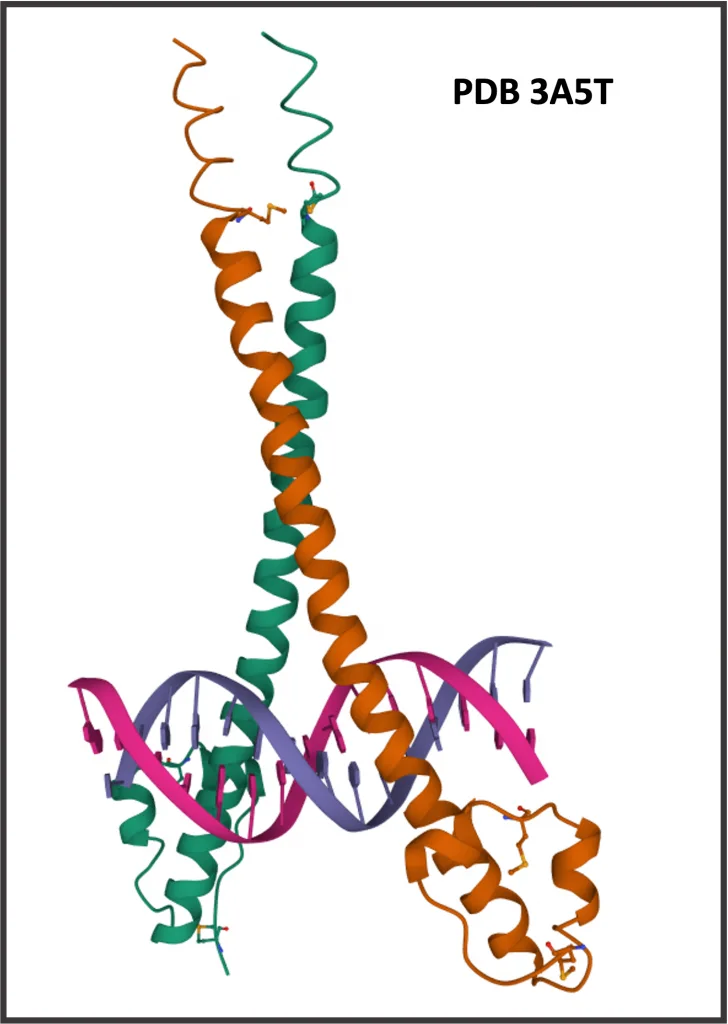
NRF2 (NFE2L2) binds DNA as a homodimer or heterodimer with small MAF factors, MAFF, MAFG or MAFK. NRF2 belongs to the family of Jun-related factors, and small MAF factors belong to the family of Maf-related factors. Both Jun-related and Maf-related families are parts of the big class bZIP, basic leucin-zipper factors (Wingender at al., 2018; TFClass link).
In the HumanPSD database NRF2 (NFE2L2) is documented for its role in more than 30 different tumors, among them are breast, colorectal, stomach, esophageal neoplasms; hepatocellular carcinoma, non-small-cell lung carcinoma, and others. Disease similarity map for hepatocellular carcinoma can be viewed here.
NRF2 protein is expressed at quite similar levels in the majority of tissues and organs investigated, with the highest level identified in esophagus and the lowest level in pancreas and bone marrow. Expression of NRF2 in normal liver is somewhat above the average level, almost the same as in thyroid, midbrain, skin and some other organs (HumanPSD database, Human Protein Atlas v20).
Expression of MAFG in the same organs and tissues is on average about 3 times lower than that of NRF2. Especially in liver, MAFG is expressed at about 20% of the NRF2 level (HumanPSD database, Human Protein Atlas v20). This suggests MAFG as a limiting factor for the NRF2/MAFG heterodimer formation in general and especially in normal liver cells.
NRF2, in different dimeric compositions, is documented to bind to its specific sites. In the TRANSFAC 2.0 database (NRF2 Locus Report), there are eight positional weight matrices for NRF2, either built on the individual genomic binding sites or based on a ChIP-seq dataset. Two of eight TRANSFAC matrices for NRF2 are “family” matrices, where several members of both Jun-related and Maf-related families are linked to. The binding motif is TGACTCAgca, which resembles very much AP-1 motif.
NRF/MAF heterodimers are known to bind to antioxidant-responsive elements (AREs). NRF2 is known to activate transcription of several functional groups of genes. Among them are ATP-binding transmembrane transporters ABCC1, ABCC2, ABCC4, ABCG2, ABCB11; metabolic enzymes CBR1, CES1, CYP2A6, GCLC, GCLM, GLO1, GPX2, UGT1A10, ADSL, AKR1B3, NOX4; proteins involved in heme biosynthesis and iron binding FTH1, PIR, AHSP; transcription factors ATF3, ATF4, ETS1, SPI1, CEBPB, TP73 as well as some other genes (TRANSFAC database).
NRF2 is acetylated by transcription co-activator CBP/p300 at several particular lysine residues within its DNA binding domain, and this post-translational modification was shown to play a positive role in transcriptional regulation by NRF2 (TRANSPATH database).
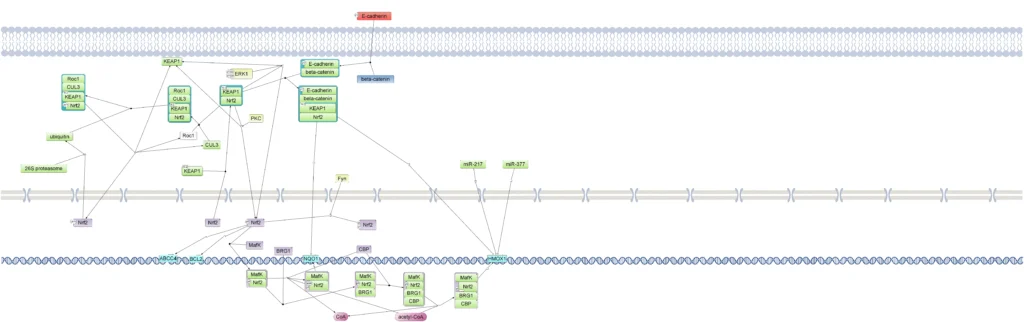
In the TRANSPATH database, there are several chains and pathways where NRF2 is involved, including Nrf2 pathway and beta-catenin pathway. Here, the diagram of Nrf2 pathway is shown (TRANSPATH database):
Many more details can be found in the integrated database TRANSFAC + TRANSPATH + HumanPSD. Here the Locus Report for human NRF2 is shown:
Open this report as a PDF file.
Pan D. et al., 2022. AKR1C3 regulated by NRF2/MAFG complex promotes proliferation via stabilizing PARP1 in hepatocellular carcinoma. Oncogene 41, 3846–3858. PMID: 35773412
Wingender E. et al., 2018. TFClass: expanding the classification of human transcription factors to their mammalian orthologs. Nucleic Acids Res. 46, D343-D347. PMID: 29087517
This page was published and last revised on 22.08.2022
Examples of some other transcription factors in cancer can be found here.
