FOXC2 in glioblastoma
TFCLass 3.3.1.3.2
FOXC2 transcription factor was recently published to bind to the promoters and upregulate expression of several genes critical for angiogenesis, ITGB3, CXCR4, and DLL4.The stability and nuclear translocation of FOXC2 was demonstrated to be maintained by the microglial exosomal circular-RNA circKIF18A (Jiang, Y. et all., 2022, Oncogene, v.41: 3461-3473). The authors demonstrated that circKIF18A can be transported from exosomes into endothelial cells and promotes angiogenesis.
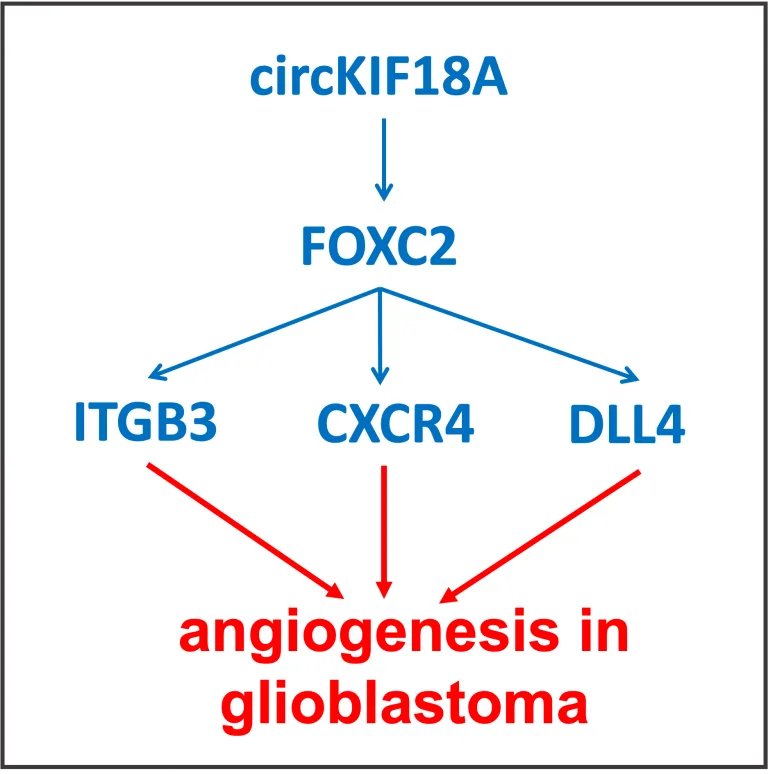
A direct binding of circKIF18A to FOXC2 was proven, and the authors even dissected the protein domain of FOXC2 involved in binding. This domain is outside of the fork head DNA-binding domain of FOXC2. Thus, FOXC2 was shown to play an important role in promoting angiogenesis in the context of glioblastoma (Jiang, Y. et all., 2022, Oncogene, v.41: 3461-3473).
FOXC2 belongs to a big family of transcription factors, Forkhead box (FOX) factors. The FOX family of factors, together with E2F-related and RFX-related factor families, constitute a class of fork head / winged helix factors, which in turn belongs to the superclass of helix-turn-helix domains (TFClass link).
FOXC2 is known to be associated with different tumors including lung, ovarian, breast neoplasms, and other tumors (HumanPSD database). In this study, the role of FOXC2 in glioblastoma was shown (Jiang, Y. et all., 2022, Oncogene, v.41: 3461-3473).
A disease similarity map for glioblastoma can be viewed here.

Expression of FOXC2 protein varies significantly between the organs and tissues investigated. The highest expression level is observed in retina followed by approximately 2-fold lower expression in medulla, kidney, heart, spinal cord, and further down to the neglectable expression in lymphocytes, monocytes, NK-cells, granulocytes and dendritic cells (HumanPSD database, Human Protein Atlas v20).
In TRANSFAC 2.0, there are 8 matrices for the FOXC2 factor. Among them, one matrix is constructed from the individual genomic binding sites and several matrices are collected from publications based on the SELEX experiments. The core binding motif for FOXC2 is GTMAATATTKAC, where M is A or C, and K is G or T (TRANSFAC database).
FOXC2 is known to be phosphorylated by ERK1 and ERK2 at multiple serine positions, and this is important for the transcriptional activation of the FOXC2 target genes (TRANSPATH database).
Many more details can be found in the integrated database TRANSFAC + TRANSPATH + HumanPSD, for instance in the Locus Report for human FOXC2:
Load failed
Open this report as a PDF file.
Jiang, Y. et all., 2022. Glioblastoma-associated microglia-derived exosomal circKIF18A promotes angiogenesis by targeting FOXC2. Oncogene, 41,3461-3473. PMID:35637250
This page was published and last revised on 19.09.2022
Examples of some other transcription factors in cancer can be found here.
