BRENDA
The Comprehensive Enzyme Information System
Unique in quality, depth and coverage of the relevant contents.
Coverage of the relevant contents.
BRENDA is the most comprehensive information repository on enzymes and enzyme ligand data. The BRENDA enzyme information system has developed into an elaborate system of enzyme and enzyme-ligand information obtained from different sources, combined with flexible query systems and evaluation tools. BRENDA has been developed and is maintained by the Institute of Biochemistry and Bioinformatics at the Technical University of Braunschweig, Germany. Data on enzyme function are extracted manually from primary literature, and are complemented by text and data mining, data integration, and prediction algorithms.
The curation process has been designed to ensure a maximum of quality, depth and coverage of the contents of this unique database. Formal and consistency checks are done by an elaborate computational pipeline. All enzymes in BRENDA are classified according to the biochemical reaction catalyzed, and are assigned to the corresponding Enzyme Commission (EC) numbers. Reaction kinetics are described in detail. BRENDA’s intuitive user interface support a wide range of queries such as fast full text search, advanced complex queries or searching via sequence or substructure. Browsing the contents is facilitated by a Taxonomic Tree, an Enzyme class, a Genome or an Ontology explorer.
Learn more about BRENDA applications and search functions on YouTube.
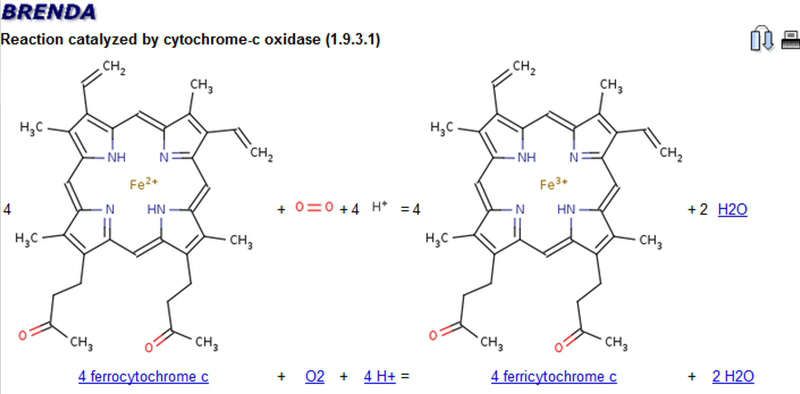
The reaction schema of the cytochrome-c oxidase catalyzed reaction.
Structure
The data sources of BRENDA comprise three main domains: Text mining data, manual annotation and external databases and ontologies.
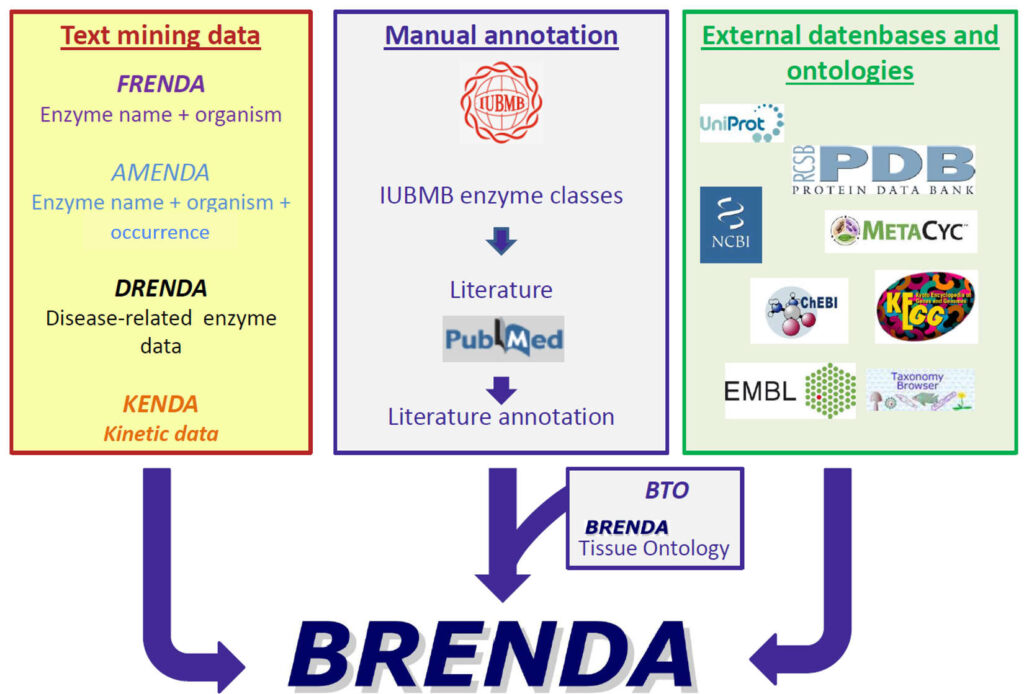
Organization of BRENDA® contents
The supplementary sources FRENDA (enzyme name & organism), AMENDA (enzyme name & organism & occurence), DRENDA (disease-related enzyme data) and KENDA (kinetic data) complement the BRENDA core by text mining data.
The manually annotated core is based on IUBMB enzyme classes and literature from PubMed. Additionally, the BRENDA tissue ontology (BTO) is linked to the manual annotation.
Cross references to several external databases like UniProt, PDB, MetaCyc, ChEBI, KEGG, EMBL, and the Taxonomy Browser of NCBI further expand BRENDA.
Key features
Enzyme and enzyme-ligand information is obtained from different sources, combined with flexible query systems and evaluation tools.
The data are acquired by manual extraction from primary literature, text and data mining, data integration, and prediction algorithms.
The manually derived core contains >3 million data points about >77,000 enzymes annotated from >150,000 publications.
BRENDA comprises molecular data from more than 30,000 organisms.
Each entry is linked to its publication source and the organism of origin. The entries are supplemented by information on occurrence, enzyme / disease relationships from text mining, sequences and 3D structures from other databases, and predicted enzyme location and genome annotation.
The human anatomy atlas CAVEman is linked to the BRENDA Tissue Ontology terms connecting functional enzyme data with their anatomical location.
Word Maps for enzymes generated from PubMed abstracts highlight application and scientific relevance of enzymes.
The EnzymeDetector genome annotation tool and the reaction database BKM-react including reactions from BRENDA, KEGG and MetaCyc.
BRENDA is the most comprehensive information repository on enzymes with 8,149 EC numbers (January 2021). Thereof 7,787 EC numbers are considered active while others are preliminary or retired and just kept for documentary purpose.
- SBML output
- Web-services
- More detailed statistics can be obtained here.
Benefits
Access the world’s largest manually curated database on enzyme data (>3 million data points annotated).
Experience BRENDA’s intuitive interface, which supports both proteomic beginners and experts in easily retrieving their data of interest.
Take advantage of all the cross-referenced other major databases like KEGG, UniProt, MetaCyc, EMBL, NCBI, and more.
You may install BRENDA on your local Linux or Windows system.
Find at your fingertips essential information from practically all fields of molecular biology, biochemistry, medicine or biotechnology.
Information downloads
BRENDA Flyer (download)
BRENDA Tutorial: Introduction (download pdf, 0.6 MB)
More BRENDA Tutorials (here)
BRENDA Exercises (download pdf, 1.7 MB)
BRENDA Video (at YouTube)
See also the BRENDA (entry at Wikipedia)
BRENDA release history (download)
How to Cite Brenda
Placzek S, Schomburg I, Chang A, Jeske L, Ulbrich M, Tillack J, Schomburg D. BRENDA in 2017: new perspectives and new tools in BRENDA. Nucleic Acids Res. 2017 Jan 4;45(D1):D380-D388. Link
Recent applications
Find below a selection of recent articles reporting about BRENDA applications.
Singh, P.K., et al. (2020) Exploring RdRp–remdesivir interactions to screen RdRp inhibitors for the management of novel coronavirus 2019-nCoV. SAR QSAR Environ. Res. 31, 857–867. PubMed
Khurshid, G., et al. (2020) A cyanobacterial photorespiratory bypass model to enhance photosynthesis by rerouting photorespiratory pathway in C3 plants. Sci. Rep. 10, 20879. PubMed
Bartman, C., et al. (2017) Factors influencing the development of visceral metastasis of breast cancer: A retrospective multi-center study. Breast 31, 66-75. PubMed
Brunk, E., et al. (2016) Systems biology of the structural proteome. BMC Syst. Biol. 10, 26. PubMed
Wei, Y., et al. (2015) Insight into Dominant Cellulolytic Bacteria from Two Biogas Digesters and Their Glycoside Hydrolase Genes. PLoS One 10, e0129921. PubMed
Tagore, S., et al. (2014) Analyzing methods for path mining with applications in metabolomics. Gene 534, 125-138. PubMed
Mayer, G., et al. (2014) Controlled vocabularies and ontologies in proteomics: Overview, principles and practice. Biochim. Biophys. Acta 1844, 98-107. PubMed
Ranjan, S., et al. (2013) Computational approach for enzymes present in Capsicum annuum: A review. Int. J. Drug Dev. & Res. 5, 88-97. Link
New release
Price request BRENDA
Publications
Schomburg I., Jeske L., Ulbrich M., Placzek S., Chang A., Schomburg D. (2017) The BRENDA enzyme information system – From a database to an expert system. J. Biotechnol. 261:194-206 PubMed
Placzek S., Schomburg I., Chang A., Jeske L., Ulbrich M., Tillack J., Schomburg D. (2017) BRENDA in 2017: new perspectives and new tools in BRENDA. Nucleic Acids Res. 45:D380-D388. Oxford.
Chang A., Schomburg I., Placzek S., Jeske L., Ulbrich M., Xiao M., Sensen C.W., Schomburg D. (2015) BRENDA in 2015: exciting developments in its 25th year of existence. Nucleic Acids Res. 43:D439-D446. Oxford.
Schomburg I., Chang A., Placzek S., Söhngen C., Rother M., Lang M., Munaretto C., Ulas S., Stelzer M., Grote A. Scheer M., Schomburg D. (2013) BRENDA in 2013: integrated reactions, kinetic data, enzyme function data, improved disease classification: new options and contents in BRENDA. Nucleic Acids Res. 41:764-772. PubMed.
Gremse M., Chang A., Schomburg I., Grote A., Scheer M., Ebeling C., Schomburg D.(2011) The BRENDA Tissue Ontology (BTO): the first all-integrating ontology of all organisms for enzyme sources. Nucleic Acids Res. 39:D507-D513. PubMed.
Barthelmes J., Ebeling C., Chang A., Schomburg I., Schomburg D. (2007) BRENDA, AMENDA and FRENDA: the enzyme information system in 2007. Nucleic Acids Res. 35:D511-D514 PubMed.
Schomburg I., Chang A., Hofmann O., Ebeling C., Ehrentreich F., Schomburg D., (2002) BRENDA: a resource for enzyme data and metabolic information. Trends Biochem. Sci. 27:54-56. PubMed
Schomburg, I., Chang, A., Schomburg, D., (2002) BRENDA, enzyme data and metabolic information. Nucleic Acids Res. 30:47-49. PubMed
Schomburg, D., Schomburg, I. (2001) Springer Handbook of Enzymes. 2nd Ed. Springer, Heidelberg Springer
Schomburg, I., Hofmann, O., Baensch, C., Chang, A., Schomburg, D., (2000) Enzyme data and metabolic information: BRENDA, a resource for research in biology, biochemistry, and medicine. Gene Funct. Dis. 3-4:109-18.
