TRANSFAC database
It’s been almost 35 years since the date of the first TRANSFAC® database publication, and since then this biggest collection of transcription factors and their genomics binding sites has served the scientific community as the most reliable and comprehensive resource for gene regulation studies.
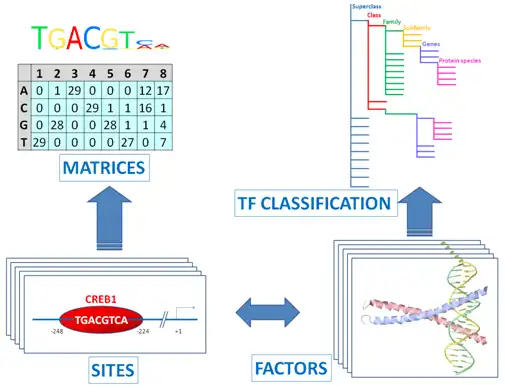
The core of TRANSFAC® comprises contents of two domains: One documents TF binding sites, usually in promoters or enhancers. The other describes the binding proteins (TFs).
Transcription factors are subsumed to classes, based on the general properties of their DNA-binding domains. This early attempt has been expanded to a comprehensive TF classification, the latest version of which can be found here.
Here is a general introduction to the online TRANSFAC® database. It shows how to operate in the basic interface and perform searches in the TRANSFAC® database.)
TRANSFAC® includes information on over 49,000 transcription factors and their genomics binding site models (PWMs – positional weight matrices) for a variety of eukaryotic species.
Binding sites referring to the same TF are merged into a positional weight matrix. Such a matrix reflects the frequency with which each nucleotide is found in each position of this TF’s binding sites and, thus, the base preference in each position.
