Pathway Omics Suite
The Pathway Omics Suite is an automated analysis pipeline that transforms transcriptomics, epigenomics, proteomics, metabolomics, and genomics datasets into a comprehensive mechanistic report.
Designed for biologists, translational researchers, and drug-discovery teams, it reveals master regulators—the key molecules that drive disease-specific regulatory cascades.
Why Researchers Choose the Pathway Omics Suite
1. Identify true mechanistic drivers—not just correlations
Uncover transcription factors, cooperative enhancers, causal pathways, and master regulators that explain why genes change and how disease mechanisms unfold.
2. Transform raw multi-omics data into actionable insights—automatically
From upload to final mechanistic report, the Suite executes a fully automated, reproducible workflow without coding or bioinformatics expertise.
3. Accelerate drug-target and biomarker discovery with explainable biology
Built on TRANSFAC® and TRANSPATH®, the system pinpoints high-value intervention points and regulatory nodes, enabling confident decisions in target prioritization and experimental design.
How Cancer Cells Outsmart Therapy: HER2+ Breast Cancer Trastuzumab Resistance in Pathway Omics Suite
Sequence and Pathway analysis (demo report)
How It Works
1. Upload & Annotate Your Data
Start with any combination of human omics data. The wizard walks you step-by-step through project creation and annotation. Supported inputs include:
- Transcriptomics: tables, FASTQ, Affymetrix, Illumina, Agilent
- Epigenomics: FASTQ, BAM, tracks, or CpG tables
- Proteomics: quantitative or list-based
- Genomics: VCF, SNP lists, FASTQ
- Metabolomics: raw tables or numerical comparisons
The suite automatically performs QC, normalization (e.g., Limma, EdgeR, Quantile), ChIP-seq peak calling, SNP lifting, enhancer mapping, and background set construction as appropriate.
2. Construct the Gene Set Driving the Biological Process
Using established statistical workflows (Limma, EdgeR, Fold Change, clustering, variance filtering), the system extracts differentially expressed genes and functionally classifies them.

3. Identify Regulatory Regions & Condition-Specific Enhancers
Regulatory regions (promoters and enhancers) are identified using CAGE-based FANTOM5 TSS annotations, Epigenomic peaks when available, Sliding-window mutation hotspot detection for genomic variants. This ensures that TF binding predictions are made on biologically relevant cis-regulatory regions.
4. Discover Transcription Factors That Control Gene Expression
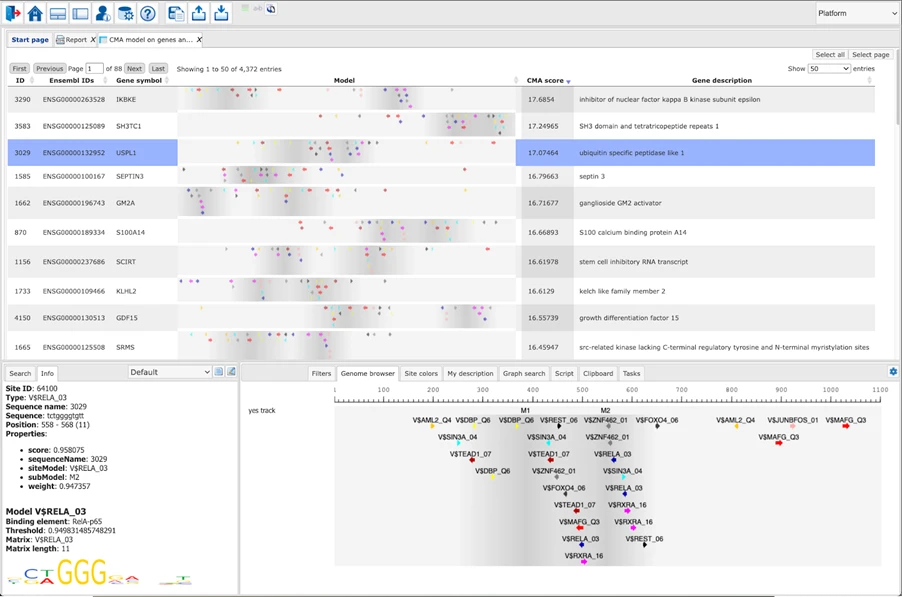
Using TRANSFAC®, Match®, and Composite Module Analyst (CMA), the suite reveals:
- TF binding site enrichment
- Cooperative TF modules.
- TFs with the strongest evidential support for regulating your differentially expressed genes.
5. Reconstruct Pathways & Identify Master Regulators using Upstream Analysis
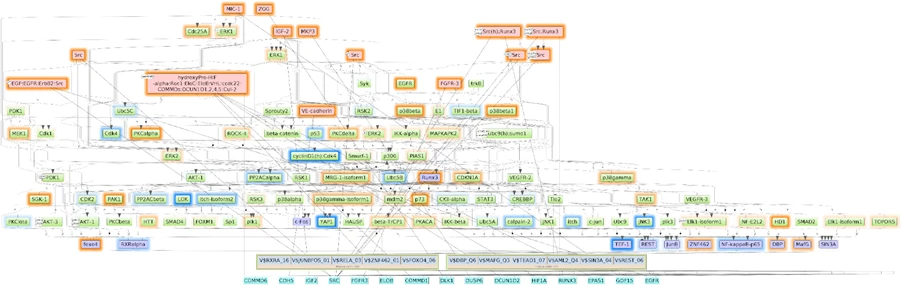
Using the TRANSPATH® signal transduction database, the software builds causal regulatory graphs upstream of key TFs. It identifies:
- Keynodes (“master regulators”)
- Intermediate molecules
- Network topology and regulatory cascade structure.
With its no‑coding GUI and robust backend (leveraging curated databases like TRANSFAC® and TRANSPATH), the Suite empowers research teams in academia and pharma to move from raw omics data to mechanistic insight — identifying key nodes in pathway regulation, actionable targets and potential biomarkers.
Key Features
End-to-end automation: From upload to full report in one workflow
Multi-omics integration: Transcriptomics, epigenomics, proteomics, metabolomics, genomics
Explainable biology: Powered by TRANSFAC® & TRANSPATH®
TFBS + enhancer modeling: Cooperative module detection via CMA
Mechanistic pathway reconstruction: Identify master regulators & druggable nodes
Publication-ready report: Structured like a scientific paper for immediate inclusion in studies
For Pharmaceutical & Biotech R&D
Designed for translational research and target discovery, Pathway Omics Suite enables pharma R&D teams to extract actionable insights from complex omics data. Through integrated regulatory and pathway network analysis, the suite identifies key upstream regulators—such as key transcription factors and master regulatory molecules—implicated in disease mechanisms.
For Academic Researchers
Built for systems biology and integrative genomics research, the Pathway Omics Suite guides scientists from raw multi‑omics data to mechanistic insight through an intuitive, end‑to‑end workflow.
What sets Pathway Omics suite apart ?
Scientifically rigorous, not a black box. Every step is transparent and based on curated databases (TRANSFAC®, TRANSPATH®) and peer-reviewed algorithms.
Mechanistic insights, not just lists. It reveals why genes change and which upstream regulators drive those changes.
Actionable for therapeutic design. By spotlighting master regulators, the pipeline directs attention to high-value intervention points.
To see how the pipeline performs in a real biomedical context, please refer to our demo report analyzing a breast cancer dataset.
