ODE modeling:
The TRANSFAC PATHWAYS package includes geneXplain platform modeling tools on the basis of BioUML simulation environment, which is according to the independent study (Maggioli F., Mancini T., Tronci E. SBML2Modelica: Integrating biochemical models within open-standard simulation ecosystems.
Bioinformatics, 2019, doi: 10.1093/bioinformatics/btz860) shown as “the best and fastest SBML simulation engine” In direct comparison to other simulation engines, such as CAPASI, SystemModeler, SMBL2Modelica and others, BioUML shows the fastest simulation time among 613 curated models of BioModels database.
Also it was shown that only BIoUML with its powerful simulation engine that supports ODE, DAE, hybrid,1D PDE passes 100% of simulation tests out of SBML Test Suite Core v3.3.0.
The BioUML simulation environment also provides a wide range of instruments for visual modeling including network modeling, composite modeling; and agent-based modeling.
BioUML web GUI for visual modeling
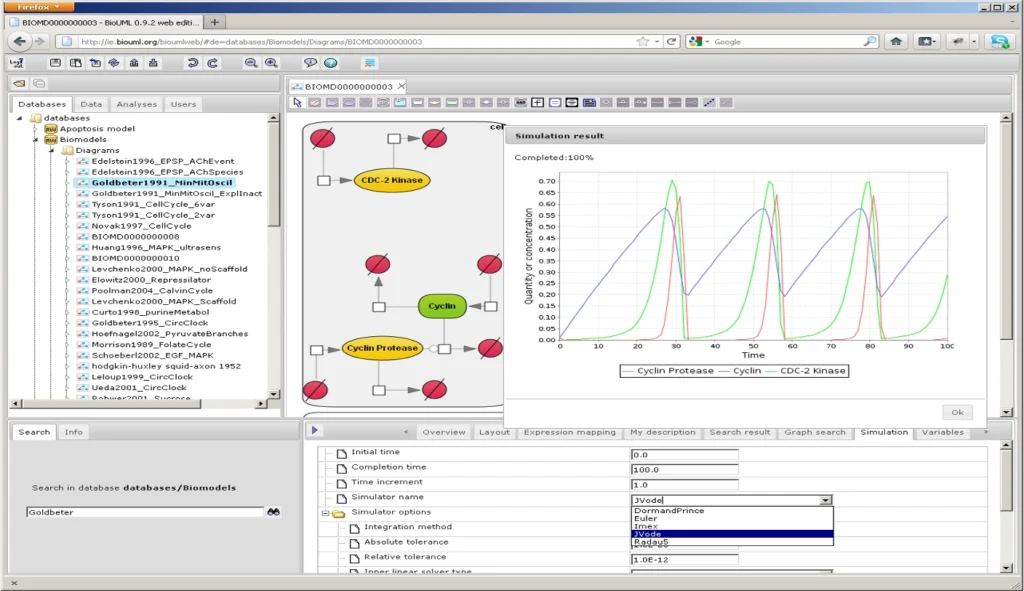
BioUML parameter fitting engine supports: fitting to time courses or steady states; multi-experiment-fitting; constraint optimization; local/global parameters; and parameters optimization using java script. The following optimization methods are implemented in BioUML: Adaptive simulated annealing, Evolutionary programming, Particle swarm, Stochastic ranking evolution strategy, and cellular genetic algorithms.
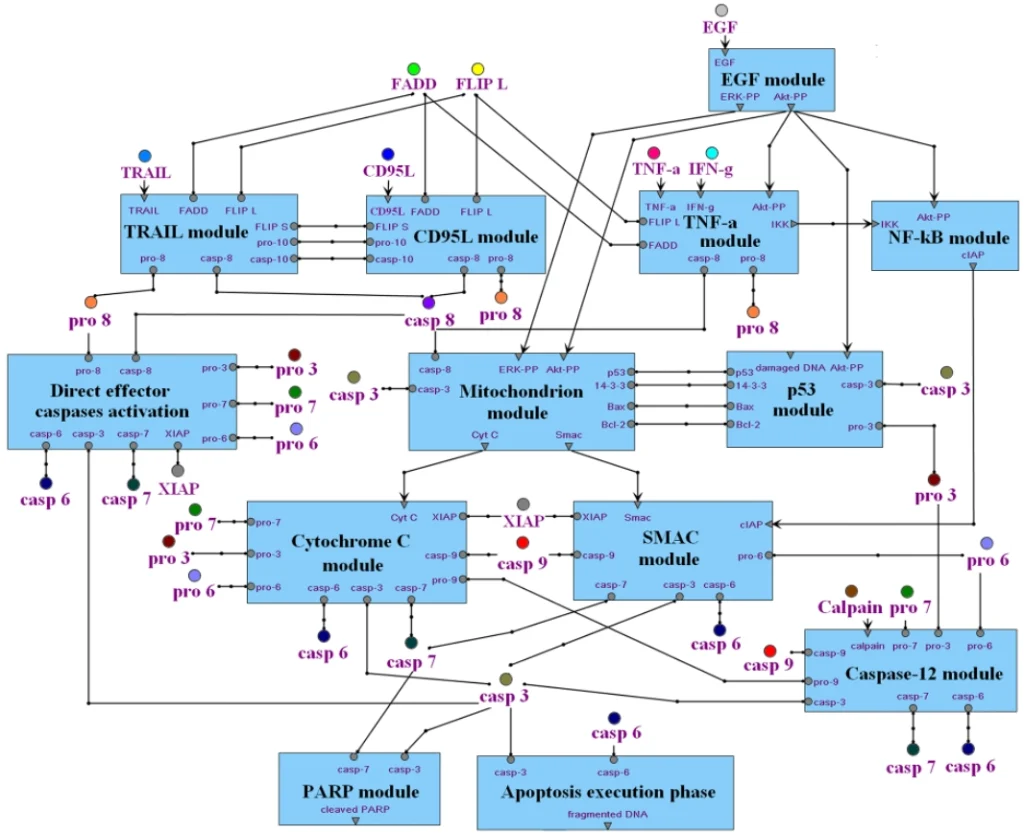
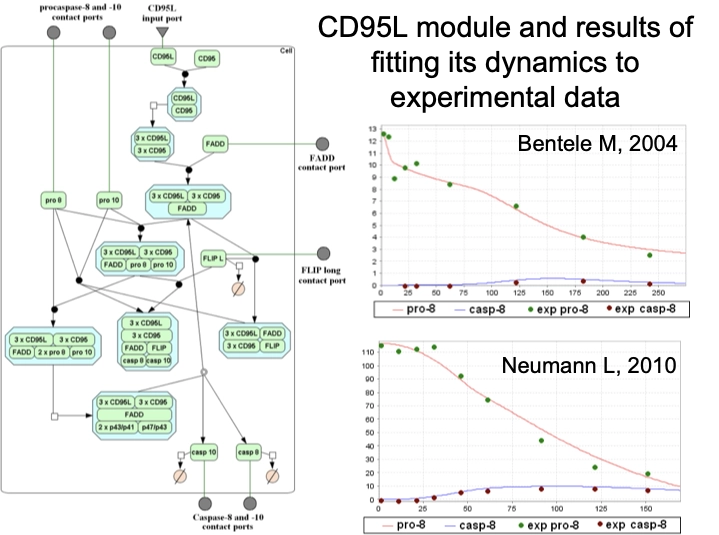
An integrated apoptosis model build using TRANSFAC PATHWAYS: