MEALR: Motif Enrichment Analysis by Logistic Regression (White Paper)
Summary: MEALR – Smarter Motif Discovery, Faster Insights
MEALR (Motif Enrichment Analysis by Logistic Regression) [1] is a next-generation tool that finds which transcription factor motifs really matter in your data — without arbitrary cutoffs or guesswork.
Instead of looking at each motif in isolation, MEALR uses sparse logistic regression to scan thousands of motifs, weigh their combined contributions, and select only those that best separate your “Yes” sequences (e.g., ChIP-seq peaks) from “No” background regions.
This reveals not just single enriched motifs but also combinatorial patterns of transcription factor binding — the real drivers of regulatory programs.
With MEALR, you can:
Integrated into the geneXplain platform, MEALR turns complex statistics into intuitive, ready-to-interpret results — making it powerful for both bioinformaticians and bench scientists.
Bottom line:
MEALR delivers a focused, high-confidence shortlist of transcription factors driving your biology, helping you move from raw data to testable hypotheses much faster.
Introduction
Motif Enrichment Analysis by Logistic Regression (MEALR) is a computational method for identifying transcription factor binding motifs that distinguish one set of DNA sequences from another.
MEALR serves as an advanced tool to uncover which transcription factor binding site (TFBS) motifs are enriched in a target sequence set (the ‘Yes’ set) compared to a background set (the ‘No’ set).
By leveraging the extensive TRANSFAC® library of position weight matrices (PWMs) and machine learning techniques, MEALR can scan thousands of candidate motifs and pinpoint a focused subset most relevant to the biological difference between the two sets.
This tool is integrated into the geneXplain platform’s user-friendly workflow environment, making it accessible to both computational bioinformaticians and bench biologists without the need for coding.
MEALR Method Overview
MEALR employs a sparse logistic regression framework to build a discriminative model that classifies sequences as Yes or No based on their motif content. Unlike traditional motif enrichment approaches that apply a strict score threshold to call binding sites, MEALR does not use any cutoff on motif scores. Instead, it computes a threshold-free sequence score for each motif in each sequence, effectively summarizing the motif’s presence/intensity across the entire sequence interval. Specifically, for each sequence x and each PWM, MEALR calculates a score equal to the log of the average of exponentiated window log-odds scores across that sequence. This ensures that all motif hits contribute, with higher-scoring hits contributing exponentially more, avoiding the need to choose an arbitrary cutoff.
The tool MEALR finds combinations of TFBS matrices that discriminate between two sets of sequences (denoted as Yes and No sets). The Yes set may consist of genomic regions identified in a ChIP-seq experiment. No sequences are often other non-coding genomic regions not overlapping with the peaks.
MEALR differs from other tools in the following points.
No cutoff or threshold is used on matrix scores to determine potential binding sites. Instead, MEALR calculates threshold-free sequence scores.
MEALR builds a discriminative model for classification which is well-established and widely applied in statistical analysis called Sparse Logistic Regression. The model consists of a linear model that estimates the probability that a sequence belongs to the Yes set based on its binding site features.
The sparseness constraint enables MEALR to select a subset of matrices relevant for classification of Yes and No sequences from a possibly large matrix library. Therefore, MEALR’s output differs from other tools by presenting a focused set of matrices.
While other site enrichment tools provided in the platform evaluate enrichment separately for each matrix, the model used in MEALR assesses the importance of matrices for discrimination in combination with other matrices of the library. Therefore, MEALR suggests (linear) combinations of transcription factor motifs.
MEALR calculates the score x of the ith sequence according to the kth matrix as
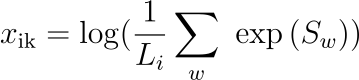
where Sw is the log-odds score of the wth window of matrix length. Each sequence is therefore associated with a vector of scores, one from each matrix, and a class (Yes, No).
Let us present an example analysis for a ChIP-seq data set consisting of 500 peak regions and 1000 sequences randomly sampled from regulatory regions across the human genome. The figure below depicts the input mask of the analysis tool.
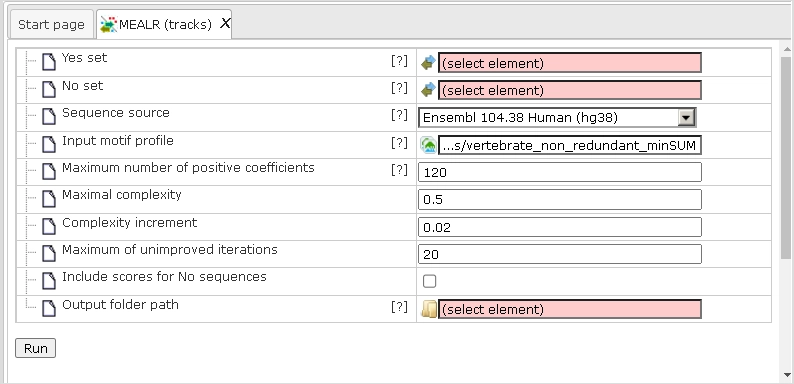
Yes set: This is the set of sequence intervals that you want to analyse, for example these can be ChIP-seq peak regions.
No set: This is the set of background intervals (control set).
Sequence source: Both Yes and No track need to refer to a common source, such as a genome, as specified by this parameter. Note that you can apply a custom source, e.g. a specifically uploaded genome. Clicking on the “Custom” option will open a new field to choose the custom sequence source.
Input motif profile: The profile lists the PWMs (motifs) that are used to assign scores to Yes and No sequences. By default, this field is set to the profile last applied in your workspace. Note that cutoffs in the profile are ignored, because MEALR calculates whole sequence scores.
Output path: In this field you select a path in the workspace to store the output table.
Output and Results Interpretation:
MEALR produces several output tables, including:
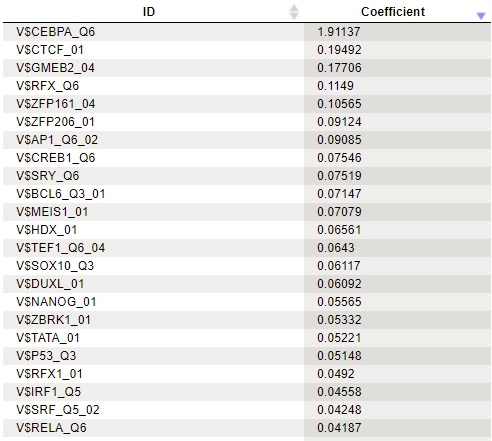
A row of the output table contains a matrix identifier and its logistic regression coefficient. The larger the coefficient value, the more important the corresponding matrix was for discriminating between Yes and No sequences. In the example above, three of the five top matrices represent members of the transcription factor subfamily C/EBP.
Applications and Use Cases
MEALR is a versatile tool in regulatory genomics for identifying key transcriptional regulators.
Across these scenarios, MEALR helps extract meaningful regulatory signals, guiding follow-up validation (e.g., ChIP or reporter assays). As part of the TRANSFAC Basic package, it links motifs to annotated transcription factors and targets, simplifying interpretation. Integrated in the geneXplain platform, it offers intuitive use for non-specialists and flexible workflows for advanced users, serving both academic and industry researchers.
Conclusion
MEALR is a state-of-the-art method for motif enrichment analysis that uses sparse logistic regression to overcome the limits of traditional tools—avoiding arbitrary thresholds, considering motif combinations, and focusing on the most predictive features. It outputs candidate motifs (and their transcription factors) that distinguish the sequence set of interest, with coefficients quantifying each motif’s importance.
This not only highlights potential regulatory drivers but also predicts regulatory potential per sequence. In practice, MEALR can both rediscover known motifs (e.g., the expected factor in ChIP-seq data) and reveal novel co-factors.
MEALR method can be accessed via geneXplain platform available in TRANSFAC Basic package.
References
1. Katie Lloyd, Stamatia Papoutsopoulou, Emily Smith, Philip Stegmaier, Francois Bergey, et al., The SysmedIBD Consortium; Using systems medicine to identify a therapeutic agent with potential for repurposing in inflammatory bowel disease. Dis Model Mech 1 November 2020; 13 (11): dmm044040. doi: https://doi.org/10.1242/dmm.044040
