Unmasking Huntington’s Disease: From a Single Gene to a Network of Pathological Signals
Huntington’s Disease (HD) remains one of the most challenging frontiers in neurology. While we have known for decades that the disease is caused by a CAG repeat expansion in the huntingtin (HTT) gene that leads to the production of mutant huntingtin protein, which ultimately causes progressive neurodegeneration. Yet, decades after this genetic cause was identified, the field continues to grapple with a fundamental question: how does a single mutation trigger such widespread and selective neuronal loss?
Recent work is beginning to close this gap by shifting attention beyond neurons to the broader cellular and regulatory landscape of the brain, where astrocytes and transcriptional control mechanisms are emerging as active drivers of disease progression rather than passive bystanders.
For researchers, the objective is clear: to identify the precise molecular “switches” that convert genetic predisposition into overt pathology — a challenge that recent advances have begun to address by leveraging the power of TRANSFAC® to bridge genomic data with disease mechanisms.
What is Huntington’s Disease?
Huntington’s is a fatal, inherited neurodegenerative disorder characterized by:
- Motor Dysfunction: Involuntary “chorea” (jerky movements) and impaired coordination.
- Cognitive Decline: Progressive loss of executive function and memory.
- Emotional Changes: Severe depression, anxiety, and personality shifts.
Pathologically, the disease is defined by pronounced degeneration of the striatum. While neuronal vulnerability has long been the focus, increasing evidence points to astrocyte dysfunction as a key contributor to the loss of neuronal homeostasis. When astrocytes lose their supportive role, they can actively promote neurodegeneration through altered signaling and extracellular matrix (ECM) remodeling.
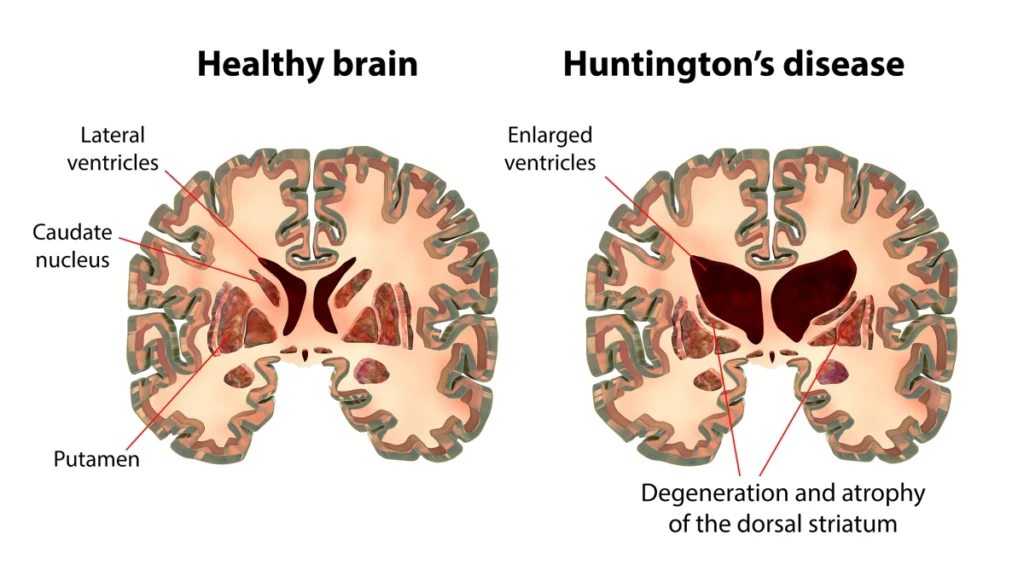
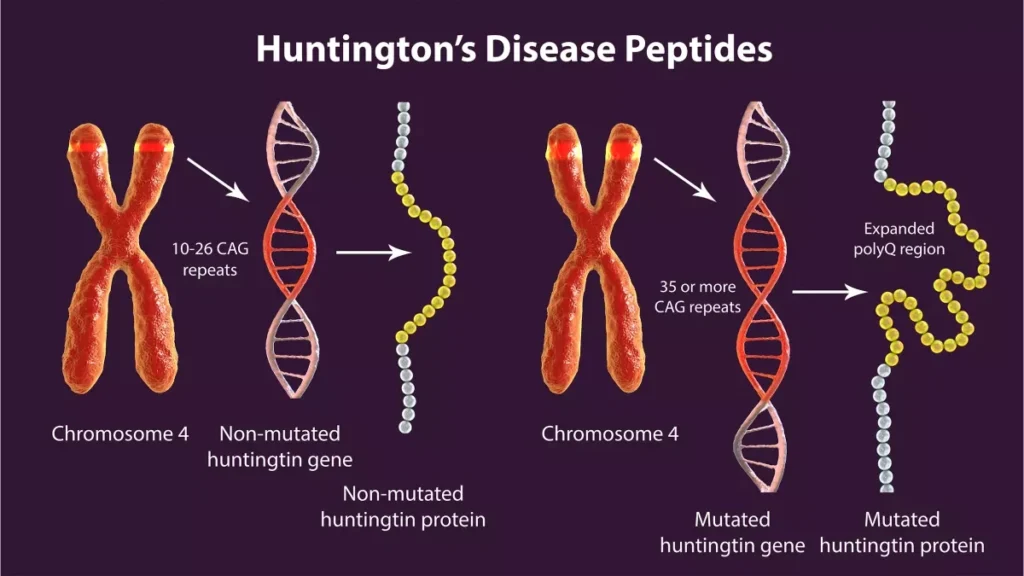
The Breakthrough Research: WNT5B and ECM Remodeling
In a new study published in Signal Transduction and Targeted Therapy (Springer Nature), researchers identified a previously unknown pathway driving the disease.
The study uncovered a pathogenic astrocyte-driven mechanism in Huntington’s disease centered on noncanonical WNT5B signaling. In HD patient tissue and mouse models, WNT5B is strongly upregulated in astrocytes, activating Ca²⁺-dependent signaling rather than canonical β-catenin pathways. Elevated WNT5B induces intracellular Ca²⁺ influx and subsequent activation of NFATc2, which translocates to the nucleus and acts as a key transcriptional regulator.
Using TRANSFAC®-driven transcription factor binding site prediction, the authors identified NFATc2 binding motifs within the MMP14 promoter (details described below). This prediction was experimentally validated, demonstrating that NFATc2 directly binds and activates MMP14 transcription, driving a matrix-degrading astrocytic phenotype. Increased MMP14 expression leads to extracellular matrix breakdown, loss of neuronal structural support, and exacerbation of striatal neurodegeneration. Together, these findings establish a linear, causative signaling axis in Huntington’s disease.
WNT5B → Ca²⁺ → NFATc2 → MMP14 → ECM degradation → neurodegeneration
Transcriptional Control of MMP14: Where TRANSFACComes In
The researchers knew that WNT5B signaling was driving the expression of MMP14, an enzyme responsible for ECM breakdown. To understand how MMP14 expression is transcriptionally regulated, the authors performed transcription factor binding site (TFBS) analysis of the MMP14 promoter. Using TRANSFAC-based positional weight matrices and promoter-scanning algorithms, they identified high-confidence NFATc2 binding motifs within ~1.5 kb upstream of the transcription start site.
By performing a bioinformatic analysis of the MMP14 promoter region, the researchers were able to:
- Map Transcription Factor Binding Sites (TFBS):
Using TRANSFAC®’s extensive library of positional weight matrices, they scanned the MMP14 promoter for potential regulators.
- Predict the Key Regulator:
The analysis identified NFATc2 as the crucial transcription factor.
- Validate Experimentally:
This bioinformatics-led hypothesis was later confirmed in the lab, proving that NFATc2 drives the pathological changes in Huntington’s Disease.
This computational prediction was not treated as an endpoint. Instead, it guided targeted experimental validation, which confirmed that NFATc2 directly binds to the MMP14 promoter and activates its transcription.
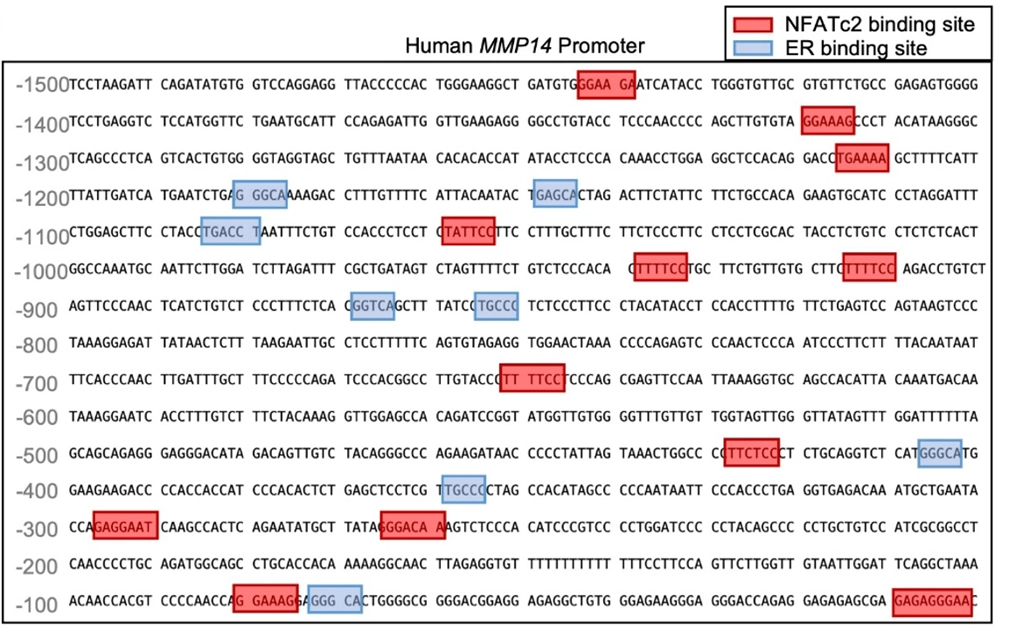
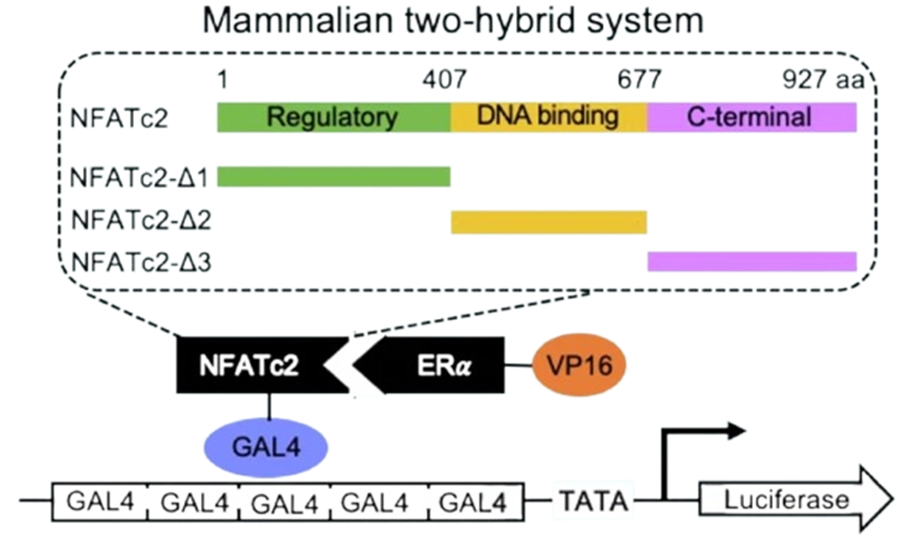
Why This Matters for Huntington’s Disease Research
This work reframes Huntington’s disease as more than a problem of mutant huntington toxicity. It highlights:
- Astrocytes as active pathological drivers, not just support cells
- Transcriptional regulation as a key amplification step between signaling and tissue damage.
- ECM remodeling as a mechanistic link between glial dysfunction and neuronal loss
Importantly, the study demonstrates how bioinformatics-guided hypotheses can dramatically narrow the experimental search space. Instead of screening dozens of candidate regulators, TFBS prediction pinpointed NFATc2 as a high-priority transcriptional switch controlling MMP14 expression.
Huntington’s Disease in a Broader Neurodegenerative Context
Huntington’s does not exist in a biological vacuum. At a molecular level, it shares significant pathways with other neurodegenerative conditions. Using a Disease Similarity Map designed via Human PSD (https://genexplain.com/human-psd-database), we can see how the gene expression signatures and regulatory networks of HD overlap with diseases like Parkinson’s, Chorea, Nervous System Diseases, Neurodegenerative Diseases etc.
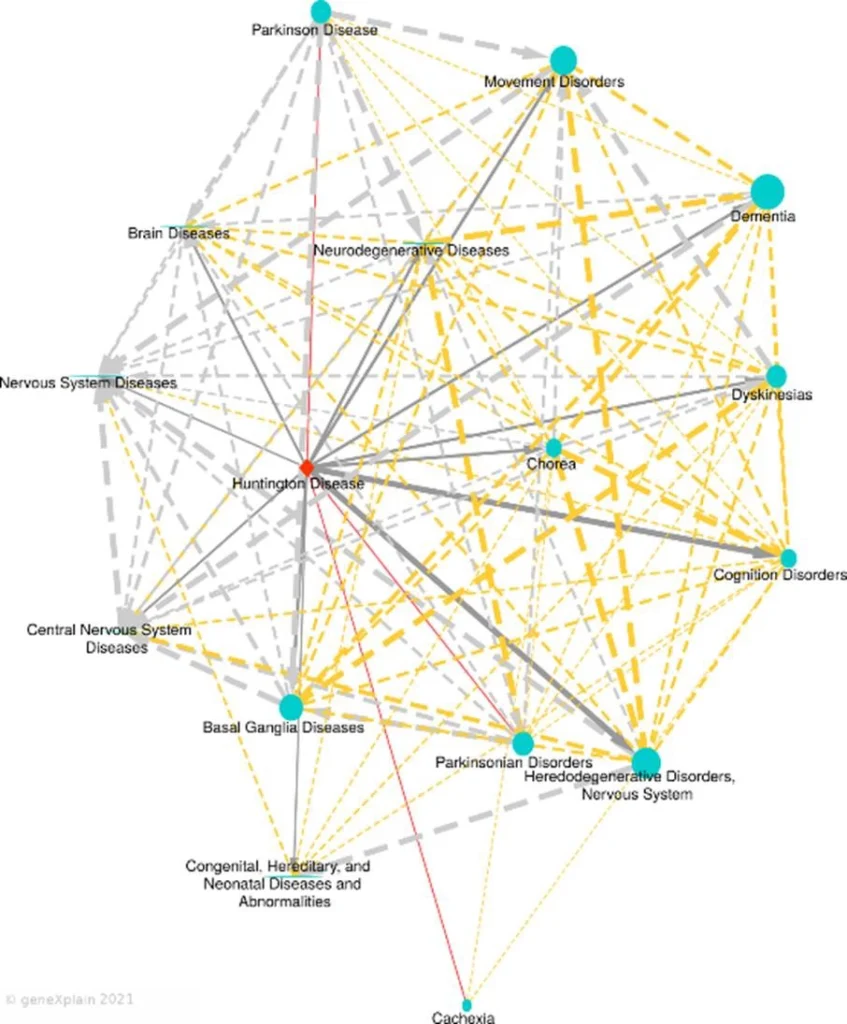
Looking Ahead
As the field moves beyond single-gene explanations, studies like this underscore the importance of integrating signaling pathways, transcriptional regulation, and cell-type-specific biology. For Huntington’s disease, understanding how astrocytes reshape the extracellular environment through transcriptionally controlled enzymes opens new directions for mechanistic research and therapeutic exploration.
The broader lesson is clear: connecting genomic variation to disease phenotype requires tools and frameworks that operate at the level of regulatory networks, not just individual genes.
References:
- Nguyen, P.T.T., Yousefian-Jazi, A., Hyeon, S.J. et al. Astrocytic noncanonical WNT5B signaling modulates extracellular matrix remodeling and neuropathology in Huntington’s disease. Sig Transduct Target Ther 11, 23 (2026). https://doi.org/10.1038/s41392-025-02545-9
- TRANSFAC®: https://genexplain.com/transfac-product/
- Human PSD: https://genexplain.com/human-psd-database/
