Glioblastoma’s vicious circle — and how we can break It
Introduction
Glioblastoma (GBM) is the most common, aggressive, and lethal form of brain cancer in adults — a Grade IV glioma that arises from the brain’s own supportive glial cells. Even when patients receive the most intensive treatment available — surgery to remove as much tumor as possible, followed by radiotherapy and the chemotherapy drug temozolomide — the outcome remains devastating.
The median survival time is only about 15 months, and nearly every case recurs. Glioblastoma infiltrates healthy brain tissue with microscopic tendrils, making complete surgical removal impossible and resistance to therapy almost inevitable.
This extraordinary resilience is not accidental.
Glioblastoma sustains itself through vicious molecular feedback loops — self-reinforcing circuits of genes and proteins that drive continuous growth and resistance. Each molecular signal amplifies the next, creating a relentless internal cycle that keeps the tumor alive, even after aggressive therapy. These loops form a hidden “engine of recurrence,” turning glioblastoma into one of medicine’s most merciless adversaries.
But even the most vicious cycle has a weak point.
Using Genome Enhancer, geneXplain’s multi-omics analysis platform, our team uncovered the key molecular “conductors” that orchestrate these loops — the master regulators that determine whether a glioblastoma remains under control or becomes unstoppable. Among them, IGFBP2, VEGFA, PDGFA, AEBP1, and OSMR stand out as the central hubs of this self-sustaining system. These molecules form positive feedback loops that continually reactivate their own signaling networks — a biochemical echo chamber that fuels tumor invasion, therapy resistance, and relapse.
Yet these loops can be broken.
By identifying and targeting the master regulators that keep them spinning, we can disrupt the tumor’s internal command system — and suggest a truly personalized approach to glioblastoma therapy.
This is not just about treating cancer; it’s about cutting the wire that keeps it alive.
Master Regulators in Glioblastoma: What Are They and Why They Matter
In complex diseases like cancer, master regulators are high-level control genes or proteins (such as transcription factors or signaling molecules) that drive numerous downstream changes, orchestrating critical tumor behaviors like proliferation, invasion, and therapy resistance in GBM. The strategic importance of these regulators lies in the idea that although GBM exhibits thousands of abnormalities, their effects converge on a smaller set of these critical “master” nodes. Identifying these key regulators provides mechanism-based biomarkers and crucial therapeutic targets—disrupting them can effectively collapse the tumor’s rogue regulatory network.
GeneXplain has developed computational tools, notably the Genome Enhancer platform, to find these master regulators from large-scale genomics data. This platform uses an automated, multi-omics pipeline combining promoter and pathway analysis. It first identifies differentially expressed genes and uses the TRANSFAC® database to scan their regulatory regions for over-represented transcription factor (TF) binding sites, thus revealing the likely controlling TFs. Next, it “walks” upstream in the signaling network using the TRANSPATH® database, searching for upstream proteins and network motifs that activate these TFs, ultimately pinpointing candidate master regulators at the top of the cascade. This data-driven, two-tiered strategy is fully automated in the Genome Enhancer pipeline, which can take raw omics data and output a report highlighting master regulators and suggesting drug candidates, making the sophisticated analysis accessible even without bioinformatics training.
Case Study: Finding Master Regulators of Poor Prognosis in GBM
To illustrate the master regulator concept, GeneXplain scientists analyzed Glioblastoma (GBM) data to understand why some patients (short-term survivors) succumb much faster than others (long-term survivors) PubMed ID (PMID) 34168757. They compared tumor samples between these groups and identified 198 genes significantly dysregulated in aggressive, short-term survivor tumors. This gene signature, which reflects aggressive GBM biology, was enriched for functions related to Epithelial-to-Mesenchymal Transition (EMT) and hypoxia response.
Upstream Analysis and Key Findings
The researchers used the Genome Enhancer pipeline to determine what was controlling these 198 genes. Promoter analysis revealed enriched binding sites for transcription factors (TFs) like NANOG, NF-κB, REST, and FRA-1 (FOSL1). Grouping these TFs showed two main modules co-regulating the tumor-promoting genes. Next, signaling network reconstruction—by looking upstream of these TFs—converged on just five key candidate master regulator proteins driving the entire dysregulated network. These were IGFBP2, VEGFA, PDGFA, AEBP1, and OSMR. Notably, these master regulators form positive feedback loops with the downstream TFs, creating a self-perpetuating circuit that maintains the aggressive state.
Validation and Impact
Among the five, IGFBP2 emerged as a central hub, regulating and being regulated by key TFs, suggesting it drives aggressiveness partly by inducing the invasion-promoting factor FRA-1. These findings were validated in independent patient cohorts, including the TCGA database, showing the master regulator genes were consistently more active in short-term survivor tumors. Crucially, high expression of IGFBP2, AEBP1, PDGFA, and OSMR correlated with significantly worse overall survival in GBM patients . This suggests these molecules are not just markers but potential drivers and therapeutic targets for breaking the tumor’s self-sustaining aggressive network. This suggests that these master regulators aren’t just markers of aggressive disease, but potentially drivers of it. They represent potential therapeutic targets – for instance, drugs that neutralize VEGF or PDGF have been clinically explored, and one could envision attempting to disrupt IGFBP2 or OSMR pathways as well to see if the tumor’s feedback loop can be broken.
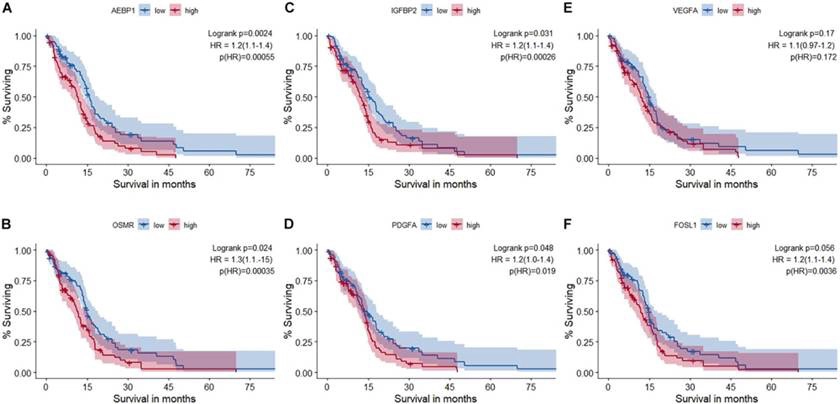
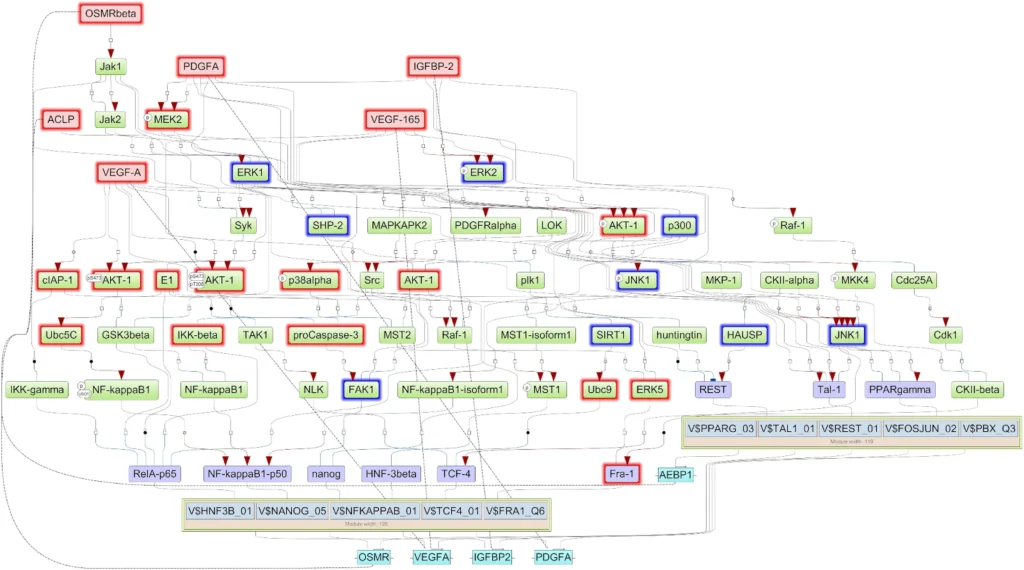
Overall, this case study demonstrates how Genome Enhancer’s upstream analysis can reveal the molecular “command and control” elements of a tumor. By going from a broad list of gene changes to a focused set of master regulators, researchers homed in on IGFBP2 and others as promising points of intervention. It’s a powerful example of turning big data into biological insight, which could eventually guide therapy (for instance, selecting patients with high IGFBP2 for trials of an IGFBP2-blocking strategy, or using AEBP1 levels as a prognostic biomarker).
Advancing Glioblastoma Research Through Collaborative Systems Medicine
The GLIOTRAIN and GLIORESOLVE projects, supported by the EU’s Horizon 2020 program, are major collaborative efforts driving new insights into glioblastoma (GBM) biology and therapy resistance. These initiatives apply systems medicine and multi-omics approaches to dissect the complex tumor microenvironment, identify key molecular regulators, and guide precision-based treatment strategies. As a core partner, geneXplain contributes its computational expertise through the Genome Enhancer platform, using advanced algorithms such as Composite Module Analyst (CMA) and upstream analysis to pinpoint GBM master regulators, model regulatory networks, and help translate multi-omics data into actionable therapeutic targets.
Conclusion and Next Steps
Glioblastoma remains one of the most challenging cancers, but the combination of cutting-edge genomics, bioinformatics, and collaborative research is shedding new light on its biology. The concept of master regulators gives us a framework to understand the disease’s complexity in a more manageable way: rather than juggling hundreds of altered genes, we target the few kingpins that govern them. GeneXplain’s Genome Enhancer platform and involvement in international projects exemplify how identifying those kingpins – and tailoring treatments to them – could be the key to finally undermining GBM’s defenses. There is still much work to be done before these insights translate to cures, but the progress is tangible. By pinpointing molecular vulnerabilities (like IGFBP2’s network or a specific TME subtype’s dependence on a pathway) and by developing tools to exploit those vulnerabilities, researchers are laying the groundwork for precision medicine in glioblastoma.
For patients and clinicians, these advances offer hope that in the future a diagnosis of GBM won’t come with such limited options. Instead of one-size-fits-all treatment, a patient’s tumor might be analyzed by platforms like Genome Enhancer to find its unique master regulators and subtype, and a personalized therapy (or combination of therapies) could be chosen accordingly. It’s an ambitious vision, but given the stakes, an essential one.
To learn more about glioblastoma and see how researchers are fighting this disease, check out this short video on glioblastoma and the latest in GBM research:
Keep an eye on our blog for future updates as we continue to unravel the secrets of glioblastoma’s master regulators and move toward more effective treatments.
References
Kalya, M., Kel, A., Wlochowitz, D., Wingender, E., & Beißbarth, T. (2021). IGFBP2 is a potential master regulator driving the dysregulated gene network responsible for short survival in glioblastoma multiforme. Frontiers in Genetics, 12, 670240.
