Core differences between canonical and non-canonical NF-κB activation pathways.
Often researches can hear about “NF-κB pathway”. Do you know about canonical and non-canonical NF-κB pathways?
1. Different cellular surface receptors.
Canonical pathway might be initiated by the following ligands via their receptors:
Non-canonical pathway might be initiated by the following ligands via their receptors:
– RANKL via its receptor RANK,
2. Differences in the intermediate signalling molecules.
A significant difference between canonical and non-canonical NF-κB pathways is in the subunit composition of IKK (Inhibitor of NF-κB Kinase) and involvement or non-involvement of IkB inhibitor.
In the canonical pathway IKK consists of two homologous catalytic subunits, IKK-a and IKK-b, and a regulatory subunit, IKK-g, also known as NEMO. Activated IKK induces the phosphorylation of NF-κB inhibitory protein IκB. Phosphorylated inhibitor IκB is then ubiquitinated and subsequently degraded, which results in the release of NF-kB dimers.
In the non-canonical pathway, IKK is a homodimer of IKK-a, and it is activated upon phosphorylation by NIK (NF-κB inducing kinase).
Activated IKK-a phosphorylates and thus activates NF-kB dimers.
3. Differences in the NF-κB subunit composition.
NF-κB family of transcription factors includes several members encoded by different genes. NFKB1 gene encodes p105 protein, which is processed to NF-κB-p50. NFKB2 gene encodes p100 protein, which is processed to NF-κB-p52. RelA gene encodes p65. Further, there are genes RelB and RelC. An active transcription factor is a heterodimer composed of two subunits.
Canonical NF-κB pathway results in activation of p50:p65(RELA) heterodimer.
Non-canonical NF-κB pathway leads to activation of p52:RELB.
All details of the canonical and non-canonical NF-κB pathways are available in the TRANSFAC Pathways or TRANSFAC Disease editions, including reactions in the format of chemical equations, chains, back references as well as pathway visualization.
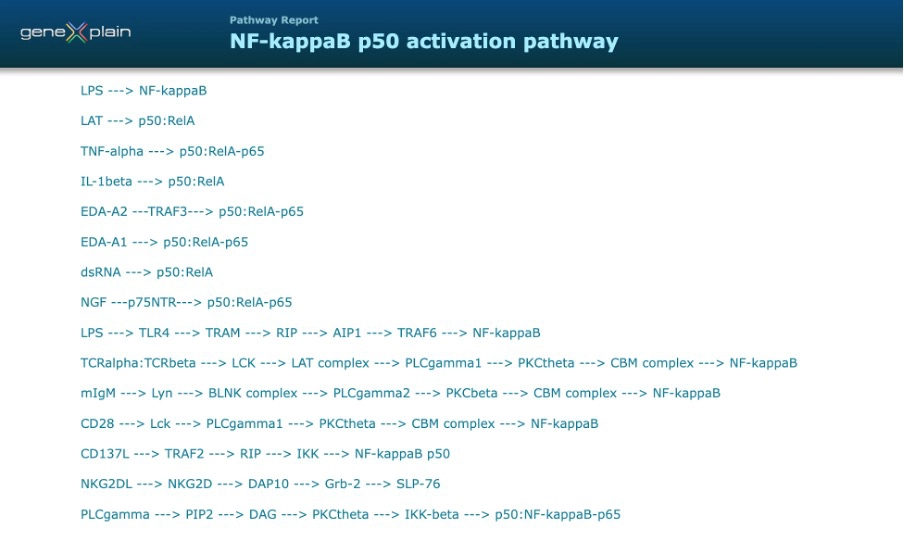
Canonical NF-κB pathway, named as “NF-kappaB p50 activation pathway” presents the following chains in the TRANSFAC Pathways or TRANSFAC Disease editions.
Non-canonical NF-κB pathway, named as “NF-kappaB p52 activation pathway” presents the following chains in the TRANSFAC Pathways or TRANSFAC Disease editions.

With our next email we provide you with the details on one canonical and one non-canonical NF-kB chain.
You are very welcome to specify what chain might be in interest, simply reply to this email.
Review of NIK (MAP3K14) central role in the non-canonical NF-kappaB pathway
Ligands and receptors of the non-canonical NF-kappaB pathway.
The non-canonical NF-kappaB pathway might be initiated by several different ligands, the members of Tumor Necrosis Factor Superfamily (TNFSF), including the following examples:
Components and mechanisms of the non-canonical NF-kappaB pathway.
NIK (MAP3K14) is NF-kappaB inducing kinase, which phosphorylates IKK-alpha (Inhibitor of NF-kappaB Kinase).
In non-active state, before receptor activation by the ligand binding, NIK is in a complex with adaptor proteins TRAF2, TRAF3, and for some of the mentioned receptors TRAF6, as well as with cIAP1/2 proteins. Protein cIAP1/2 provides proteasomal degradation of NIK minimizing the amount of this kinase.
Upon ligand binding, these receptors recruit TRAF adaptors and cIAP1/2, which prevents NIK ubiquitination and degradation, causing rapid NIK accumulation and thus activation.
Activated NIK phosphorylates IKK-alpha and facilitates its binding to NFKB2/p100.
Within a protein complex with NIK, IKK-alpha phosphorylates NFKB2/p100, which is then ubiquitinated and processed into NF-kappaB p52.
NF-kappaB p52 heterodimerizes with another member of the NF-kappaB family of transcription factors, RelB. Heterodimers p52:RelB are translocated into the nucleus where they are involved in transcriptional regulation of their specific target genes.
Tissue-, organ- and cell-specific expression of the receptors.
The receptors of the non-canonical NF-kappaB pathway are characterized by a tissue-enriched expression, that is their expression is significantly higher in several organs, tissue- or cell-types as compared to other studied tissues. Tissue specificity of the receptors defines a location and physiological function of a particular non-canonical chain leading to p52:RelB activation. Here, details of tissue-specific expression of several TNF receptor superfamily members are shown.
LT-betaR (TNFRSF3) expression is significantly enriched in liver, pancreas, mammary gland.
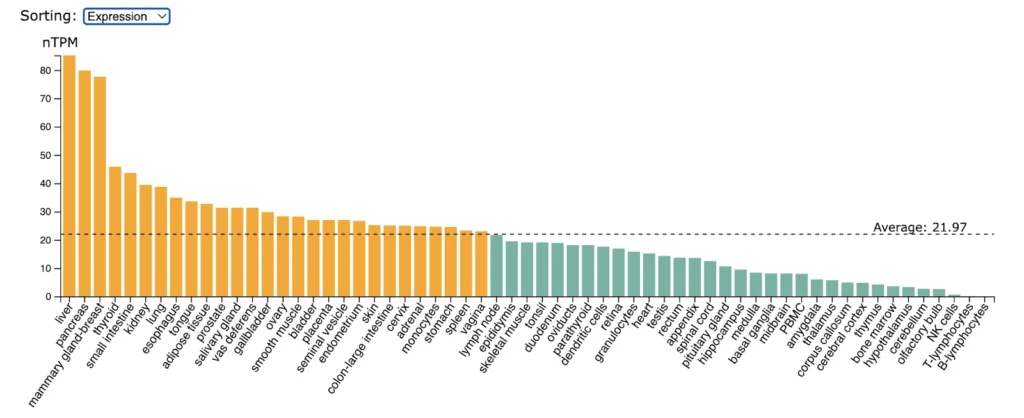
CD30 (TNFRSF8) expression is significantly enriched in monocytes followed by lower but still enriched expression in basal ganglia, adipose tissue, appendix, granulocytes, stomach.
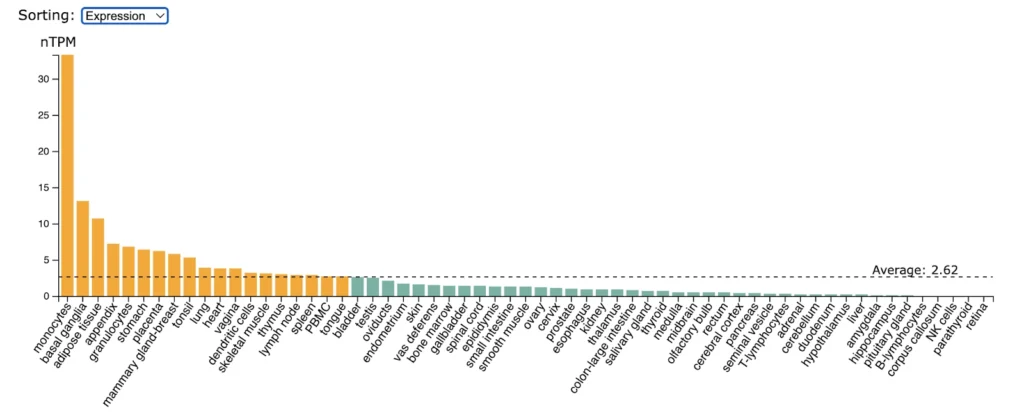
RANK (TNFRSF11A) expression is significantly enriched in salivary gland, gallbladder, small intestine, duodenum, colon, rectum.
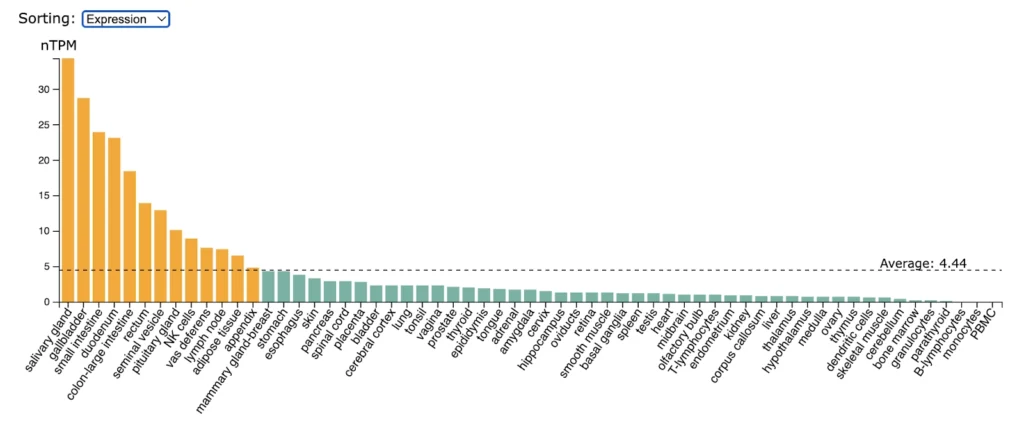
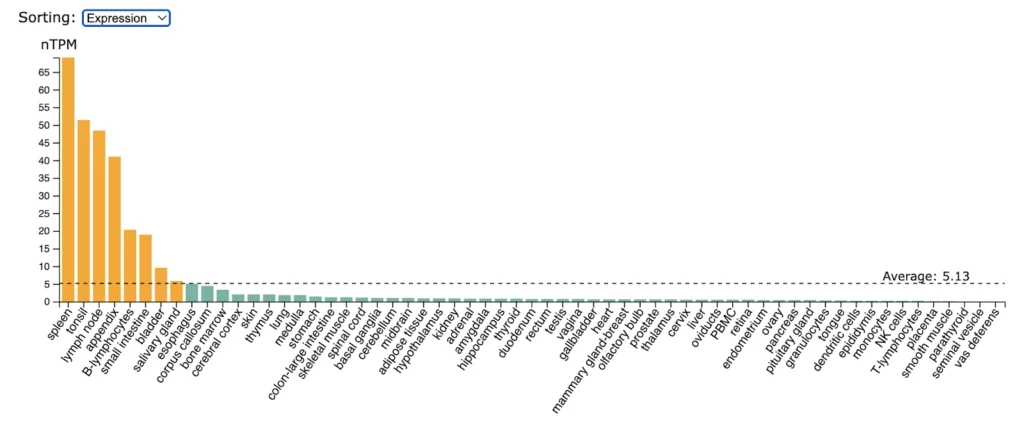
BAFFR (TNFRSF13C) expression is higly enriched in spleen, tonsil, lymp nodes, appendix, B-lymphocytes, small intestine.
All the details of the non-canonical NF-kappaB pathway are available in the TRANSFAC Pathways or TRANSFAC Disease editions, including reactions in the format of chemical equations, expression & disease involvement for each component, back references as well as pathway/chain visualization.
RANKL —> RANK —> NIK —> IKK-alpha —> NF-kappaB2-p52:RelB (chain)
https://portal.genexplain.com/cgi-bin/build_hpt/idb/1.0/pageview.cgi?view=PathwayReport&chain_acc=CH000006751
CD154 —> CD40 —> NIK —> IKK-alpha —> NF-kappaB2-p52:RelB (chain)
https://portal.genexplain.com/cgi-bin/build_hpt/idb/1.0/pageview.cgi?view=PathwayReport&chain_acc=CH000006755
(LT-alpha:LT-beta) —> LT-betaR —> NIK —> IKK-alpha —> NF-kappaB2-p52:RelB (chain)
https://portal.genexplain.com/cgi-bin/build_hpt/idb/1.0/pageview.cgi?view=PathwayReport&chain_acc=CH000006756
LIGHT —> LT-betaR —> NIK —> IKK-alpha —> NF-kappaB2-p52:RelB (chain)
https://portal.genexplain.com/cgi-bin/build_hpt/idb/1.0/pageview.cgi?view=PathwayReport&chain_acc=CH000006757
THANK —> BAFFR —> NIK —> IKK-alpha —> NF-kappaB2-p52:RelB (chain)
https://portal.genexplain.com/cgi-bin/build_hpt/idb/1.0/pageview.cgi?view=PathwayReport&chain_acc=CH000006759
CD70 —> CD27 —> NIK —> IKK-alpha —> NF-kappaB2-p52:RelB (chain)
https://portal.genexplain.com/cgi-bin/build_hpt/idb/1.0/pageview.cgi?view=PathwayReport&chain_acc=CH000006808
CD30L —> CD30 —> NIK —> IKK-alpha —> NF-kappaB2-p52:RelB (chain)
https://portal.genexplain.com/cgi-bin/build_hpt/idb/1.0/pageview.cgi?view=PathwayReport&chain_acc=CH000006821
