From Regulatory Networks to Longevity: A Genome Enhancer Case Study
Introduction
In our previous exploration, we framed aging as a breakdown of gene regulatory networks and highlighted transcription factors like FOXO3 as master regulators of exceptional longevity. We also suggested that systems biology could reveal upstream “command centers” of aging.
Here, we extend that approach using the Genome Enhancer platform to analyze human longevity-associated variants. Focusing on lifespan rather than specific diseases, we identified key transcription factors—SATB1, YY1, Fra-1 and HNF4A—along with the master regulators, FOXO3, PRKG1 and EPHA6, that form central signaling nodes.
In this post, we examine how these regulators connect through signaling pathways, why upstream analysis is essential for uncovering causal mechanisms in complex traits like aging, and how network-based insights can inform drug repurposing strategies—including candidates such as Flavopiridol and alpha-lipoic acid.
Mapping Longevity Networks with Genome Enhancer
To probe the genetic underpinnings of human longevity, we turned to a specialized analysis pipeline – Genome Enhancer. This platform was fed a list of genomic loci (SNPs) statistically associated with longevity from https://www.longevitygenomics.org/downloads.
The analysis proceeded in two major steps:
- Transcription Factor (TF) Identification: First, Genome Enhancer scanned the genes affected by these longevity SNPs (e.g. genes harboring the variants or in their vicinity) to find transcription factors that consistently regulate those genes. In other words, which TFs might “sense” the DNA variations and alter gene expression in long-lived individuals?
- Upstream Master Regulator Search: Next, the software “walked” upstream through known signal transduction networks (using the comprehensive TRANSPATH database) to find higher-level regulators that control the activity of those TFs. Think of it as tracing the wiring diagram upstream from the transcription factors to see what kinases, receptors, or signaling molecules feed into them.
This two-tiered approach – from SNPs, to affected genes, to key TFs, to master regulators – exemplifies the power of upstream analysis in complex traits. Rather than looking at one gene at a time, Genome Enhancer reconstructed a regulatory network architecture for longevity.
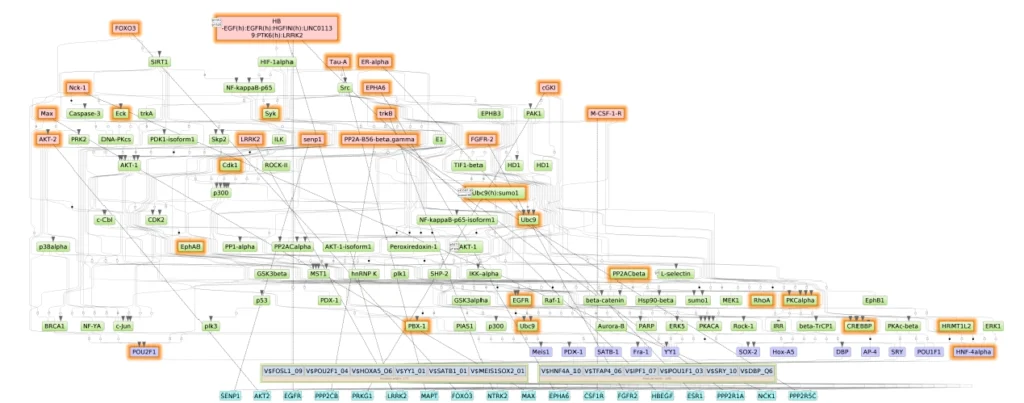
The result is a system-level view: transcription factors at the DNA control layer, embedded within larger signaling pathways regulated by master nodes. Figure 1 below illustrates a portion of this network, showing how the identified master regulators and TFs are connected via intermediate signaling molecules in cells.The master regulators FOXO3, EPHA6 and PRKG1 (highlighted in red) sit at the top, feeding into pathways (green nodes) that ultimately modulate the activities of SATB1, YY1, Fra-1 and HNF4A (blue nodes) on longevity genes. This kind of network map is not just a pretty picture – it provides testable hypotheses for intervention. If we can tweak one of the red nodes, we might recalibrate an entire ensemble of downstream longevity processes.
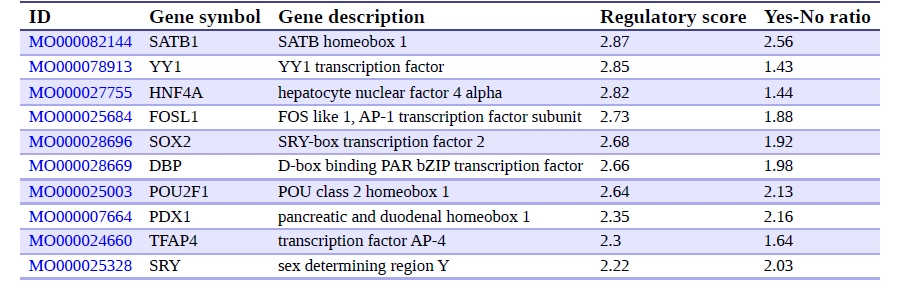
Critical Transcription Factors: SATB1, YY1, FOSL1 and HNF4A
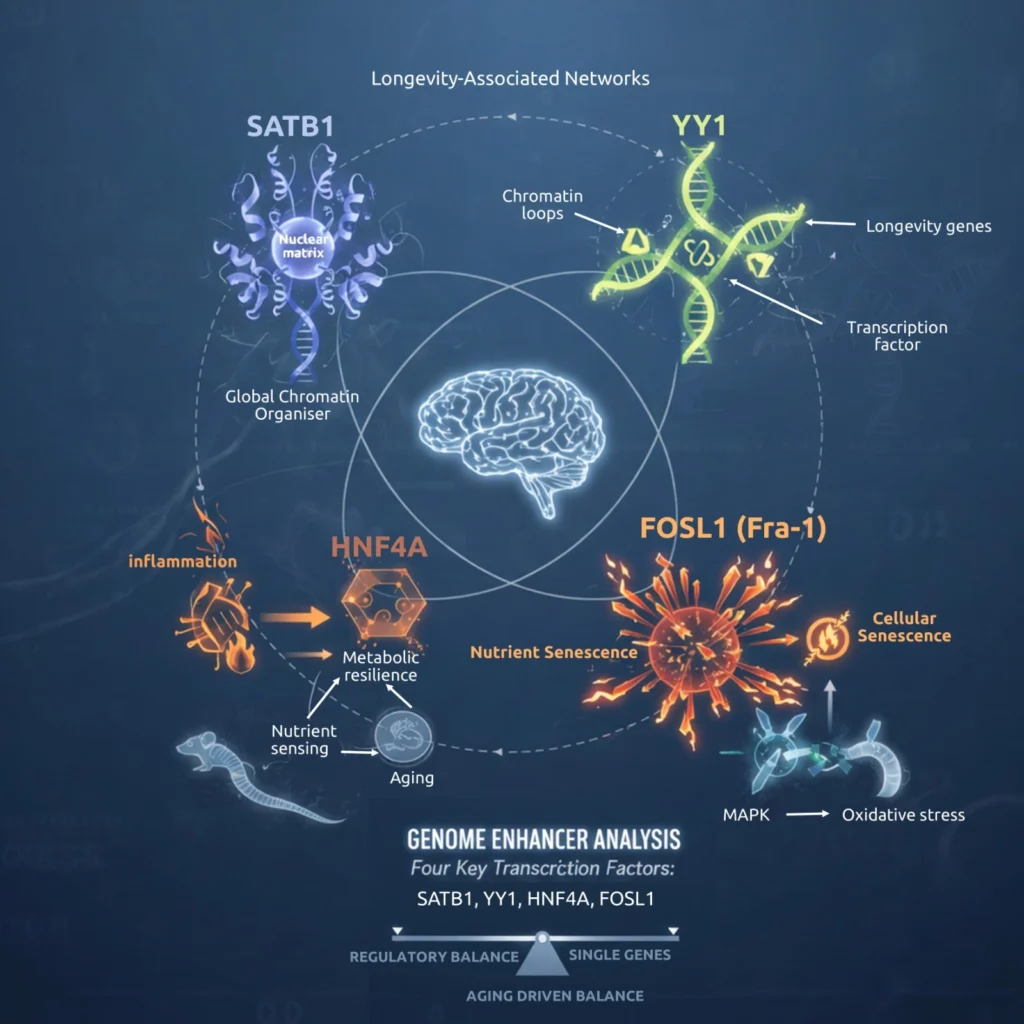
Genome Enhancer identified SATB1, YY1, HNF4A, and FOSL1 (Fra-1) as central transcriptional regulators in longevity networks, demonstrating that lifespan is shaped not by a single gene but by key regulatory nodes that control numerous downstream targets. Misregulation of just a few of these hubs can mimic cellular aging, whereas stabilizing their coordinated activity—balancing chromatin integrity, metabolic resilience, and stress-inflammatory signaling—may help delay or mitigate hallmarks of aging.
- SATB1 – SATB1 (Special AT-rich Sequence Binding Protein 1) is a global chromatin organizer that anchors DNA to the nuclear matrix and regulates higher-order chromatin loops. Genome Enhancer identified SATB1 as a regulator of multiple longevity-associated genes. In mice, SATB1 expression in the hypothalamus correlates positively with lifespan and declines with age and metabolic disease. Experimental loss of SATB1 induces p21 activation and premature cellular senescence, suggesting it protects against age-associated dysfunction by maintaining chromatin stability. Thus, higher SATB1 activity may support a youthful transcriptional program, whereas its decline may promote genomic dysregulation and aging phenotypes.
- YY1 (Yin Yang 1) is a multifunctional transcription factor involved in enhancer–promoter looping and epigenetic regulation. Genome Enhancer identified YY1 as a regulator of longevity-associated genes, consistent with evidence that YY1 binding declines in aged stem cells, weakening chromatin loops and driving age-related transcriptional drift. Knockdown of YY1 in young cells induces aging-like gene expression patterns. Together, these findings position YY1 as a genomic stabilizer whose sustained activity may help preserve transcriptional control and delay aging.
- HNF4A (Hepatocyte Nuclear Factor 4-alpha) is a nuclear hormone receptor that regulates glucose and lipid metabolism and functions downstream of insulin/IGF-1 signaling. Although not classically defined as a longevity gene, HNF4-related factors are required for dietary-restriction–mediated lifespan extension in model organisms, including C. elegans (NHR-62). HNF4A expression declines with age in metabolic tissues, correlating with epigenetic silencing and dysfunction, and it can induce senescence programs in certain contexts. These findings position HNF4A as a key link between metabolic resilience, nutrient sensing, and aging—consistent with the lifespan-extending effects of caloric restriction.
- FOSL-1: FOSL1 (Fra-1) is a stress-responsive component of the AP-1 transcription factor complex that promotes proliferation, inflammation, and tissue remodeling—processes that often become dysregulated with aging. Activated downstream of MAPK and oxidative stress pathways, AP-1/Fra-1 activity is associated with cellular senescence and pro-inflammatory gene programs, including SASP-like signatures. Because AP-1 factors can compete with protective regulators such as FOXO proteins at shared enhancers, the balance between Fra-1–driven inflammatory signaling and stress-resilience programs may influence aging trajectories. Thus, FOSL1 represents a key regulatory node linking kinase signaling, chronic inflammation, and age-related transcriptional reprogramming.
Finding the Command Centers of Aging: Upstream Analysis
Identifying key transcription factors is only part of the story. The more fundamental question is: what regulates those regulators? This is where upstream master regulators become critical. Read more about upstream analysis here:
https://genexplain.com/upstream-analysis-identifying-master-switches-in-gene-regulation/
Even without any prior information about aging biology, the system independently pinpointed FOXO3 as a master regulator within the longevity-associated dataset. This identification was entirely data-driven—the algorithm inferred FOXO3’s central role solely from network relationships derived from SNP-associated gene regulation patterns, without any built-in bias toward known aging genes.
This is biologically compelling. FOXO3 is one of the most consistently replicated longevity-associated genes in human studies. It activates programs central to lifespan regulation, including:
- Metabolic adaptation
- Autophagy
- DNA repair
- Oxidative stress resistance
All of these processes are tightly linked to enhanced cellular resilience and extended lifespan. Lot of literature already provides evidence on the role of FOXO3 with longevity We have discussed FOXO3’s central regulatory role in aging in detail in our blog post https://genexplain.com/foxo3-key-regulator-in-aging-and-longevity/
A Regulatory Tug-of-War: FOXO3 vs Fra-1
As mentioned above Genome Enhancer detected enriched motifs for AP-1 transcription factors, particularly Fra-1 (FOSL1). This is not random. Emerging research suggests that FOXO3 and AP-1 represent opposing functional programs:
- FOXO3 promotes cellular protection, repair, stress resistance, and metabolic stability.
- Fra-1/AP-1, activated by stress-associated kinases (e.g., MAPKs), drives proliferation, inflammation, and tissue remodeling.
What makes this especially fascinating is where this opposition occurs.
FOXO3 and Fra-1 do not merely regulate separate gene sets. They frequently compete for regulatory control at shared enhancers and promoters, often using overlapping chromatin regions and co-factors. In other words, they participate in a molecular “rope-pulling” mechanism at critical regulatory nodes.
Genome Enhancer revealed that many longevity-associated SNPs alter transcription factor binding sites precisely within these shared regions. This suggests that genetic variants in long-lived individuals may subtly shift regulatory balance:
- Favoring FOXO3-driven resilience
- Over AP-1-driven inflammatory or proliferative stress programs
Longevity, therefore, may not be encoded in a single gene. Instead, it emerges from how regulatory balance is tuned at critical control points — determining whether stress adaptation or inflammatory remodeling dominates gene expression programs.
Genome Enhancer also identified two signaling-level master regulators: PRKG1 and EPHA6. These molecules sit above the transcriptional layer and connect extracellular signals to genome-wide regulatory programs.
PRKG1 (Protein Kinase G Type I) is a cGMP-dependent kinase involved in nitric oxide signaling and neuroendocrine regulation. Network analysis places it upstream of key longevity transcription factors. Notably, a 2024 study showed that selective PRKG1 knockdown in hypothalamic neurons extended lifespan and improved metabolic resilience in mice[23], validating it as a functional lifespan regulator rather than a statistical artifact. Because cGMP/PKG modulators already exist in cardiovascular medicine, PRKG1 represents a pharmacologically tractable intervention node in aging.
EPHA6 (Eph Receptor A6) is a receptor tyrosine kinase involved in neuronal development and signaling and has emerged in human longevity GWAS studies. As a signaling hub capable of activating MAPK and PI3K/Akt pathways, EPHA6 sits upstream of longevity-associated transcription factors and may influence systemic aging through neural circuits. Given that tyrosine kinases are a well-established druggable class, EPHA6 is a promising candidate for therapeutic exploration in longevity biology.
A Coherent Regulatory Architecture
Together, FOXO3, AP-1/Fra-1, SATB1, YY1, HNF4A, PRKG1, and EPHA6 form a multi-layered regulatory system:
- Genetic variants alter TF binding at shared regulatory regions
- Transcription factors compete for control of stress-response programs
- Upstream kinases and receptors modulate transcriptional balance
- System-level signaling determines cellular aging trajectories
This systems-level architecture reinforces a central theme:
Aging is not driven by a single “longevity gene,” but by interconnected signaling and transcriptional networks whose balance determines resilience or decline.
From Networks to Therapeutics: Drug Repurposing for Healthy Aging
Mapping longevity networks enables identification of compounds that modulate key master regulators such as PRKG1 and EPHA6. Using HumanPSD knowledge and in silico predictions, Genome Enhancer generated candidate drugs targeting these nodes, highlighting two illustrative examples.
Flavopiridol (Alvocidib), a CDK inhibitor developed for cancer, reshapes transcriptional programs by targeting CDK9 and CDK1/2. Beyond tumor suppression, CDK inhibition may reduce senescence-associated p16/p21 activity and dampen pro-inflammatory SASP signaling. Flavopiridol has shown cognitive benefits in Alzheimer’s models[33], suggesting potential to counter “inflammaging.” While oncology use is limited by toxicity, controlled dosing strategies could make it relevant for senescence or immune modulation.
Alpha-Lipoic Acid (ALA), a mitochondrial cofactor and antioxidant, emerged due to predicted effects on metabolic and stress-response pathways. ALA supports mitochondrial function, activates NRF2, elevates glutathione, and mitigates oxidative stress—factors relevant to PRKG1 and HNF4A signaling. Although lifespan effects are mixed, ALA improves metabolic and cognitive parameters in aging models and is clinically well tolerated.Together, these examples illustrate network-driven drug repurposing: instead of screening thousands of molecules blindly, targeting upstream regulators offers rational, mechanistically grounded intervention strategies. While experimental validation is essential, this systems-based approach narrows the search space and accelerates translation toward geroprotective therapies.
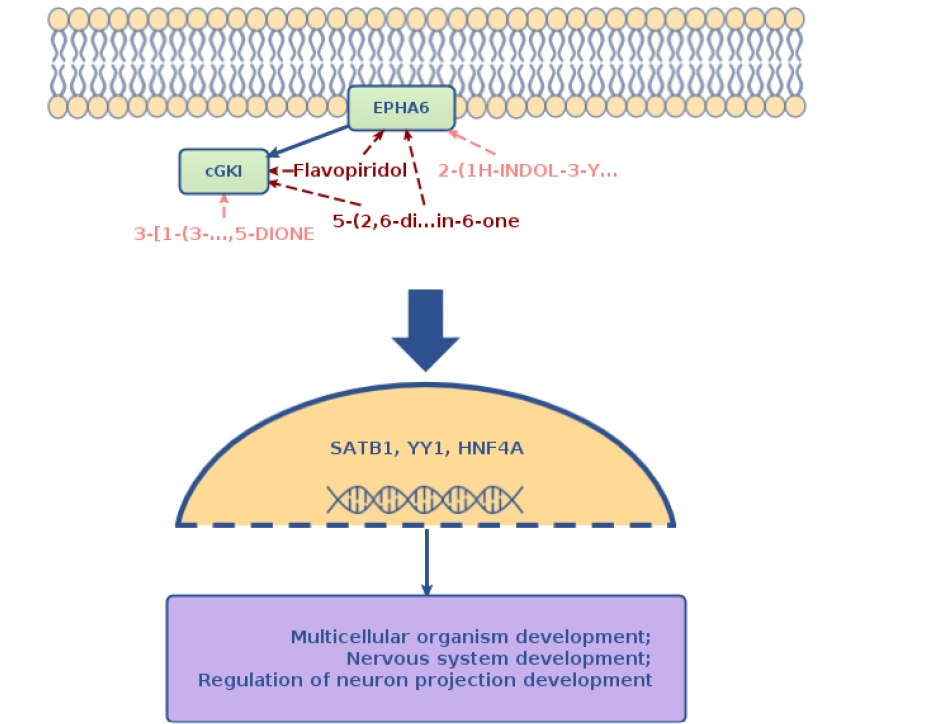
Conclusion: From Variants to Regulatory Control
Lifespan emerges from complex interactions among metabolic, inflammatory, neuroendocrine, and stress-response pathways—meaning that focusing on isolated genes often misses the underlying regulatory logic. Using a systems biology approach, Genome Enhancer traced diverse longevity-associated variants across DNA repair, immune, and mitochondrial genes back to shared transcriptional regulators (SATB1, YY1, HNF4A, Fra-1) and upstream signaling hubs (FOXO3, PRKG1, EPHA6).
This upstream convergence shows how distinct genetic variants can influence common master switches, producing similar longevity outcomes. By identifying these high-level controllers, network analysis shifts aging research from descriptive omics toward causal mechanisms and actionable intervention points within the human aging interactome.
References:
- Willcox BJ et al. PNAS. 2008. Association of FOXO3A genotype with human longevity.
https://www.pnas.org/doi/10.1073/pnas.0801030105 - Flachsbart F et al. PNAS. 2009. Confirmation of FOXO3A association with longevity.
https://www.pnas.org/doi/10.1073/pnas.0902410106 - Greer EL et al. PLOS Biology. 2010. Role of CBP and SATB-1 in Aging, Dietary Restriction, and Insulin-Like Signaling.
https://journals.plos.org/plosbiology/article?id=10.1371/journal.pbio.1000245 - Liu Z et al. Cell Stem Cell. 2019. Loss of SATB1 Induces p21-Dependent Cellular Senescence.
https://www.cell.com/cell-stem-cell/fulltext/S1934-5909(19)30348-0 - Zhang W et al. Nature Communications. 2024. Disruption of YY1-mediated super-enhancer–promoter looping drives transcriptomic changes during mammalian stem-cell aging.
https://pmc.ncbi.nlm.nih.gov/articles/PMC12155207/ - Deng Z et al. Journal of Biological Chemistry. 2008. YY1 restrained cell senescence through repressing the transcription of p16.
https://pubmed.ncbi.nlm.nih.gov/18558095/ - Hou L et al. Ageing Research Reviews. 2018. Emerging topics in C. elegans aging research.
https://www.sciencedirect.com/science/article/pii/S0047637418300836 - Thakur A et al. Aging (Albany NY). 2010. Maternal diet, aging and diabetes meet at a chromatin loop.
https://www.aging-us.com/article/100330/text - Ning BF et al. Oncogene. 2019. Nuclear receptor HNF4α performs a tumor suppressor function.
https://www.nature.com/articles/s41388-019-1080-3 - Fight Aging! 2024. A Subset of Cells in the Hypothalamus Regulates Longevity in Mice.
https://www.fightaging.org/archives/2024/11/a-subset-of-cells-in-the-hypothalamus-regulates-longevity-in-mice/ - Broer L et al. Scientific Reports. 2016. Novel loci and pathways significantly associated with longevity.
https://www.nature.com/articles/srep21243 - Dines M et al. Neuroscience Letters. 2008. Learning and memory impairment in Eph receptor A6 knockout mice.
https://www.sciencedirect.com/science/article/abs/pii/S0304394008004722 - Aging and Senescence Review. PMC. 2020. Senescence as an Amyloid Cascade: The Amyloid Senescence Hypothesis.
https://pmc.ncbi.nlm.nih.gov/articles/PMC7248249/ - Shay KP et al.Southern Medical Journal. 2009. Can α-Lipoic Acid Mitigate Progression of Aging-Related Decline Caused by Oxidative Stress?
- https://sma.org/southern-medical-journal/article/can-%CE%B1-lipoic-acid-mitigate-progression-of-aging-related-decline-caused-by-oxidative-stress/
