End‑to‑end multi‑omics workflow – upload, integrate, and analyse genomics, transcriptomics, epigenomics, proteomics and metabolomics
Modern omics technologies routinely deliver long lists of differentially expressed genes, methylation changes, or protein abundance shifts. However, these downstream readouts do not immediately explain which signaling cascades are responsible for the observed changes, or which upstream regulators should be targeted to modulate the system.
The upstream of TFs concept closes this gap. Starting from transcription factors (TFs) inferred from promoter/enhancer analysis, we trace the regulatory architecture backwards through signaling networks to the receptors, kinases, and other key nodes that drive these TFs. This reveals causal links between:
This “from TFs upstream to signaling” strategy is at the core of geneXplain’s pathway-centric analysis and provides a powerful bridge between regulatory genomics and systems-level signaling biology.
From gene expression changes to signaling cascades
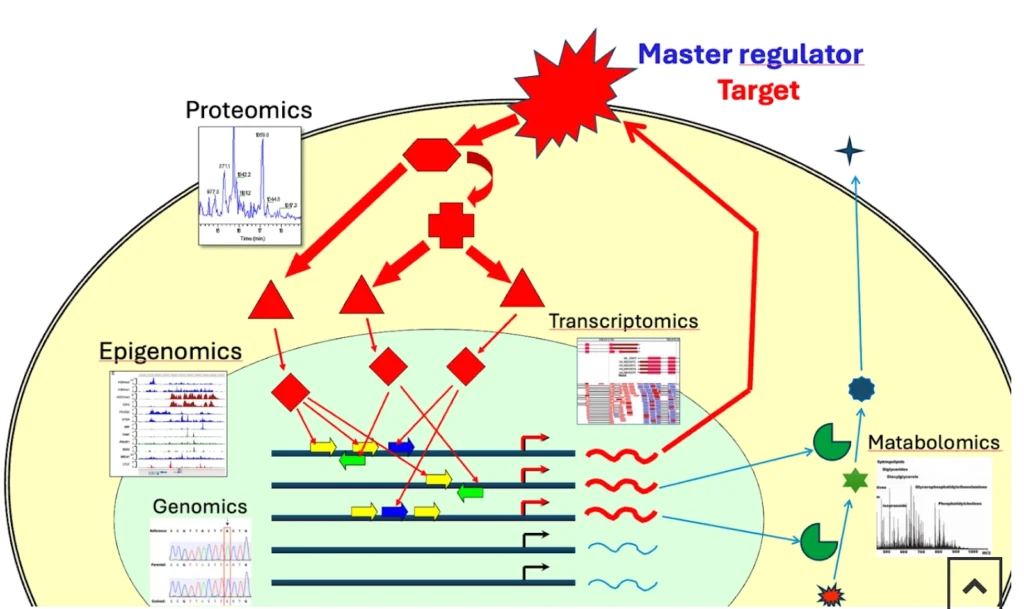
Typical omics workflows start with one or more of the following layers:
By integrating these layers, we obtain a robust gene set characterizing the condition of interest. The key question then becomes:
Which transcription factors and upstream signaling cascades are responsible for this gene set?
Answering this question requires connecting regulatory regions (where TFs act) with signaling pathways (where extracellular and intracellular signals are processed).
Key steps in upstream-of-TF analysis
Step 1 – Constructing the gene set describing the process
The analysis begins by constructing a gene set that best represents the studied biological process or pathology:
The result is a set (or several sets) of genes that show coordinated behavior in the condition under study and serve as the entry point for regulatory and pathway analysis.
Step 2 – Promoter/enhancer analysis and TF identification
For each gene set, promoter and/or enhancer regions are analyzed to detect overrepresented transcription factor binding sites (TFBSs):
This step transforms a “flat” gene list into a regulatory profile: a small set of transcription factors and TF modules that are statistically most likely to control the observed gene expression changes.
Step 3 – Mapping TFs onto signaling networks
Once candidate TFs are identified, the analysis proceeds upstream into signaling space:
This step builds signal transduction routes that explain how external or internal cues propagate through the network and converge on the TFs controlling the gene set.
Step 4 – Upstream network reconstruction and master regulators
The next goal is to identify master regulators – those key upstream nodes whose activity can influence large parts of the network:
These master regulators provide a mechanistic explanation of the phenotype and serve as high-value hypotheses for further experimental validation.
Step 5 – Pathway and process interpretation
With TFs and upstream regulators defined, the analysis moves to biological interpretation:
This provides a systems-level view that connects:
- the original omics readouts (e.g. DEGs),
- the TFs inferred from regulatory region analysis, and
- the upstream signaling modules that explain those TF activities.
Multi-omics context for upstream-of-TF analysis
The upstream-of-TF method becomes particularly powerful when combined with multiple omics layers:
By integrating these data types, we not only predict which TFs and upstream nodes could be important, but also obtain independent evidence that they are active and relevant in the studied system.
What you gain from upstream-of-TF analysis
Applying the upstream-of-TF approach allows you to:
This upstream-of-TF strategy is implemented in geneXplain’s analysis environment, where promoter/enhancer analysis, TF motif enrichment, and signaling-network reconstruction are integrated into a coherent workflow, providing a clear mechanistic bridge from transcription factors to signaling cascades.
