Finished Projects
PD-MitoQUANT

A quantitative approach towards the characterisation of mitochondrial dysfunction in Parkinson’s disease


Mitochondrial dysfunction is implicated in Parkinson’s Disease (PD), but a detailed understanding of the cause and effect in αSyn toxicity is lacking. Through the provision of quantitative and systematic characterization of mitochondrial dysfunction, PD-MitoQUANT will provide an unprecedented understanding of the role of mitochondrial dysfunction in PD, identify and validate novel disease biomarkers, and propose innovative therapeutic targets that can be further progressed by the EFPIA partners. The consortium leverages multi-disciplinary expertise in the fields of αSyn biochemistry, iPSC-derived PD models, mitochondrial function and structural analysis, proteotoxicity, ER stress and UPR signaling, systems biology of mitochondrial function, and in vivo animal models. A key focus will be quantitative description and integrated analysis of mitochondrial function and its relation to proteotoxicity; representing a key panel of consortium partners Prehn [RCSI], Abramov [UCL], Corti [ICM] and Koopmann [RUMC] who also have assembled long-standing expertise in primary neuron culture, iPSC-derived neurons, PD in vivo models, proteostasis and ageing studies.
These investigations will be supported by expert teams in iPSC-derived in vitro PD models and in vivo PD models from the academic partners (Hunot [ICM], Melki [CNRS], Di Monte [DZNE], Broccoli [CNR], SME [Mimetas] and EFPIA partners [Teva], [Lundbeck] and [UCB]. Through integrated in-vitro, in-silico and in-vivo approaches, and supported computationally by SME [GENEXPLAIN], PD-MitoQUANT will perform thorough and unprecedented investigations of mitochondrial dysfunction.
Finally, the consortium will initiate a European research platform of excellence investigating mitochondrial dysfunction in PD continuing beyond the project, further supported by [PUK]’s PD human tissue biobank. This will provide long-term and sustainable progress in the understanding of mitochondrial dysfunction in PD and towards clinical application.
It is geneXplain’s role in this consortium to apply its proprietary methodology of computational upstream analysis and master regulator search to the data generated by the project partners.
miRNA-DisEASY

microRNA biomarkers in an innovative biophotonic sensor kit for high-specific diagnosis
This collaborative project is funded under Horizon 2020 – Marie Skłodowska-Curie Actions Research and Innovation Staff Exchange (MSCA-RISE) and by the Brazilian National Council for Scientific and Technological Development (CNPq).
Since the discovery of human microRNAs (miRNAs) in 2001, progress in genomics and transcriptomics has increased our comprehension of the role of miRNAs in gene expression regulation. MiRNAs are small, noncoding RNA molecules that are promising to serve as clinical biomarkers and diagnostic tools for illness and disease, as significant changes in their expression occur in response to pathological states. However, miRNA detection is still challenging nowadays, since costly, complex sample preparations and RNA amplifications are not yet reliable enough for clinical decision making. In this context, three innovative EU companies, Optoi Microelectronics (with microelectronic knowhow and manufacturing skills), DestiNA Genomics (with ‘error free’ chemical technology for microRNAs recognition) and geneXplain (with bioinformatic capabilities) joined forces with four key academic research groups from the Universities of Trento (Italy), Granada (Spain), Hannover Medical School (Germany) and Santa Catarina (Brazil). The parties combined their knowledge on miRNA biomarkers with a particular focus on lung cancer, to develop a novel and reliable miRNA detection system for this disease. To this end, research collaborations will address the analysis of miRNAs specifically over-expressed in lung cancer, continuing experimental work already started in the past years, but with greater integration and focus.
GeneXplain’s contribution to the project:
Task 1.4 Promoter and pathway analysis of miRNA profiles [GENEXPLAIN: M12-M36]. miRNA expression profiles will be analysed with promoter analysis and pathway analysis tools. Mechanism-based prioritization of miRNA differentially expressed between AD and SCC will be done.
Task 6.1 IPR [OPTOI, DESTINA and GNX; M1-M48] In compliance with the Consortium Agreement will be established a strategy framework for the treatment of IP generated within the project. Projects results will be periodically reviewed, assessed and reported to implement the strategy for protection and/or publication.
Task 6.2 Exploitation plan [OPTOI, DESTINA and GNX] Projects results will be assessed and reported to draft a exploitation/commercialization plans & strategy, after appropriate protection where applicable.
Task 6.4 Outreach and communication [OPTOI and GNX; M1-M48] Supervision and coordination of planned dissemination and outreach activities. All partners will promote outreach events and activities to communicate the project to the public (§3.3). All seconded participants will take part to outreach activities within the hosting institution related initiatives for communication of the project mission and objectives, and, where relevant, of project outcomes.
OxidoCurin
Targets and biomarkers of the novel anticancer compounds inducing oxidative stress in cancer cells


As a Eurostars project, OxidoCurin is an international, SME-driven project for innovative product development with a fast track to market.
The project aims at further developing a novel anti-cancer lead compound, to make it suitable for application in preclinical and later clinical trials. The starting point will be a previously characterized lead compound. To provide all preconditions for future exploitation, the target(s) of the compound will be experimentally validated by different methods, among them state-of-the-art TPP (thermal proteome profiling) methodology.
Elucidating the mechanism of interaction of the lead compound with its target(s) will enable to identify its mechanism of action. Further efforts will be made to identify biomarkers of sensitivity and resistance of cancer cells and the mechanism of possible toxic side effects.
Project funding info and brief overview.
miRCol

Identification of miRNA-transcription factor network motifs to search for drug repurposing candidates in the treatment of colorectal liver metastasis


This project is funded by the German-Russian Funding Competition in the Area of Industry-Oriented Applied Research and Cooperation of Innovative SMEs 2016
Colorectal cancer (CRC) is the third most common cancer in the world with a frequently lethal complication – the development of colorectal liver metastases (CLM). Despite improved methods for detection and treatment of CLM, the chemotherapy methods used today remain inadequate and no molecularly targeted therapy is available. Therefore, this project aims at the combined use of cancer genomics with computational drug repositioning to predict treatment responses of existing drugs based on disease associated gene regulatory networks. The objectives of the project are to combine most innovative knowledge-based bioinformatic techniques built on databases and methods of artificial intelligence and modern experimental methods to analyze chromatin structure and to do massive gene expression analyses. From these data, the intracellular gene regulatory and signaling networks specific for heterogeneous forms of colorectal liver metastasis (CLM) will be reconstructed. They will be used to reveal the most important components of these network and the feedforward loops (FFLs) consisting of miRNA and TFs, to identify most sensitive nodes in these circuits for potential therapeutic intervention, and to predict the best drug combinations and treatment regimens for multi-targeted therapies of CLMs. Exploitation of the results of the project will be in patenting the drug combinations and treatment regimens for multi-targeted therapies of CLMs as well as in offering to the drug industry the developed bioinformatics network-motif-based drug target identification and drug repurposing pipeline for potential application in various cancers and other disease areas.
GeneXplain’s contribution to the project is in
- scanning available relevant data to identify miRNAs;
- determining promoter and enhancer regions for genes related to CRC / CLM;
- developing advanced algorithms to find common TF regulatory sites in miRNA and CRC / CLM genes;
- integrating these results with miRNA target predictions using bioinformatic resources;
- reconstructing combined TF and miRNA regulatory networks of CRC / CLM genes;
- establishing a reference protein-protein interaction database for CRC / CLM genes;
- finding TFs in feed-forward loops (FFL), which are also highly represented in the CRC / CLM PPI database;
- detecting possible drug attack points by modeling the FFLs;
- selecting drug candidates.
GlioTrain

Exploiting GLIOblastoma intractability to address European research TRAINing needs in translational brain tumour research, cancer systems medicine and integrative multi-omics


This collaborative project is funded under Horizon 2020 – Innovative Training Networks (ITN) under the call H2020-EU.1.3.1. – Fostering new skills by means of excellent initial training of researchers.
Glioblastoma (GBM) is the most frequent, aggressive and lethal of all brain tumours. It has a universally fatal prognosis with 85% of patients dying within two years. New treatment options and effective precision medicine therapies are urgently required. This can only be achieved by focused multi-sectoral industry-academia collaborations in newly emerging, innovative research disciplines. GLIOTRAIN will exploit the intractability of GBM to address European applied biomedical research training needs. The ETN, which comprises 9 beneficiaries and 14 partner organisations from 8 countries, will train 15 innovative, creative and entrepreneurial ESRs. The research objective of GLIOTRAIN is to identify novel therapeutic strategies for application in GBM, while implementing state of the art next generation sequencing, systems medicine and integrative multi-omics to unravel disease resistance mechanisms. Research activities incorporate applied systems medicine, integrative multi-omics leveraging state of the art platform technologies, and translational cancer biology implementing the latest clinically relevant models. The consortium brings together leading European and international academics, clinicians, private sector and not-for-profit partners across GBM fields of tumour biology, multi-omics, drug development, clinical research, bioinformatics, computational modelling and systems biology.
Thus, GLIOTRAIN will address currently unmet translational research and clinical needs in the GBM field by interrogating innovative therapeutic strategies and improving the mechanistic understanding of disease resistance. The GLIOTRAIN ETN addresses current needs in academia and the private sector for researchers that have been trained in an environment that spans translational research, medicine and computational biology, and that can navigate confidently between clinical, academic and private sector environments to progress applied research findings towards improved patient outcomes.
Using the geneXplain platform, the task of one of the trainees will be to identify combinations of transcription factors which bind to drug-resistance-specific enhancers in the genome and to reveal master regulators responsible for the dysregulation of gene expression in resistant GBM. Goals are (i) to release an open software prototype for prediction of GBM drug-resistance-specific enhancers in the genome using “upstream analysis”, (ii) to model signaling cascades activated in resistant GBM and identification of master regulators, and (iii) to identify master-regulator-based biomarkers of resistance.
ExITox-II

Explain Inhalation Toxicity-2
Animal-free mechanism-based toxicity testing – predict toxicity after repeated dose inhalation exposure by using a read across approach.


A collaborative project funded by the German Federal Ministry of Education and Research (BMBF) in the funding program
“e:ToP – Innovative Toxikologie zur Reduzierung von Tierversuchen”
ExITox-2 was built on the outcomes of ExITox-1 and aimed to develop an integrated approach for testing and assessment (IATA) to replace in vivo studies with repeated inhalation exposure. We established this IATA with the help of a read across (RAX) approach of ExITox-1, in which we tested groups of chemically similar compounds that share a specific mode of action (MoA) or adverse outcome pathway (AOP) in vivo. In ExITox-1 we showed that biological profiles based on the transcriptome changes and functional outcomes from in vitro and ex vivo models can be used to differentiate RAX groups with similar mode of action. Beside the RAX group vinyl ester from ExITox-1, we have included two new AOPs in second phase, namely pulmonary inflammation, and fibrosis. In the course of ExITox-2, geneXplain was expanded the “complete upstream analysis”, which is an integrated bioinformatics promoter analysis of the DEGs, identifying potential transcriptional regulators, with a proprietary pathway analysis controlling the activity of these transcriptional regulators and converge in defined sets of so-called master regulators (Kel et al., 2006).
Further the geneXplain platform was extended with lung-specific pathways up to newly constructed master pathways (for fibrosis and inflammation) to compare toxic effects to general systemic responses.
ExITox-2 integrated four new key aspects into the IATA:
- The uptake of compounds after inhalation exposure were estimated by physiological pharmacokinetic modeling and QSAR models.
- The onset of gene changes and miRNA regulation, by dose dependent testing, were investigated to better differentiate between group-specific changes and general stress responses related to high dosing.
- We generated and used master pathways per group to differentiate AOP related changes from general cellular stress responses.
- An IATA was developed, which allows to predict the toxicity of an untested compound in a read across context based on the generated data.
Optogenerapy

Optogenetic Protein Therapy for Multiple Sclerosis


A collaborative RIA (Research and Innovation Action) project funded by the European Union Horizon 2020 Programme (H2020) under H2020-EU.2.1.3. – INDUSTRIAL LEADERSHIP – Leadership in enabling and industrial technologies – Advanced materials.
Optogenerapy proposes a new interferon-β (IFN-β) drug delivery system to revolutionize Multiple Sclerosis treatment.
The aim is to develop and validate a new bio-electronic cell based implant device to be implanted subcutaneously providing controlled drug release during at least 6 months. The cell confinement within a chamber sealed by a porous membrane allows the device to be easily implanted or removed. At the same time, this membrane acts to prevent immune rejection and offers long-term safety in drug release while overcoming the adverse effects of current cellular therapies. Wireless powered optogenetics – light controlling the cellular response of genetically engineered cells – is used to control the production of IFN-β.
Replacing standard intravenous IFN-β delivery by subcutaneous delivery prevents short and long term side effects and efficiency-losses related to drug peaks and discontinuation, while saving non-adherence costs.It is a low-cost system enabling large scale manufacturing and reduction of time to market up to 30% compared to other cell therapies, combining:
- Polymeric biomaterials with strong optical, biocompatibility and barrier requirements, to build the cell chamber and to encapsulate the optoelectronics.
- Optoelectronics miniaturization, autonomy and optical performance.
- Optimal cellular engineering design, enhanced by computer modelling, for stability and performance of the synthetic optogenetic gene pathway over long-term implantation.
- Micro moulding enabling optoelectronics and membrane embedding for safety and minimal invasiveness.
The innovation potential is so huge that a proof-of-concept was listed by Scientist Magazine as one of the 2014’s big advances in science.
In this top-class consortium, industrial pull meets technological push, ensuring that the preclinically validated prototype obtained at the end responds to market demands.
GeneXplain’s role is to model promoters that are optimally responding to the synthetic pathways that will be generated in the project.
MyPathSem
A data integration platform for generating patient-specific signaling pathways for personalized treatment decisions in clinical applications
Eine Datenintegrationsplattform für die Generierung von Patienten-spezifischen Signalwegen für individualisierte Behandlungsentscheidungen in klinischen Anwendungen


A collaborative project funded by the German Federal Ministry of Education and Research (BMBF) in the funding program
“i:DSem – Integrative Datensemantik in der Systemmedizin”
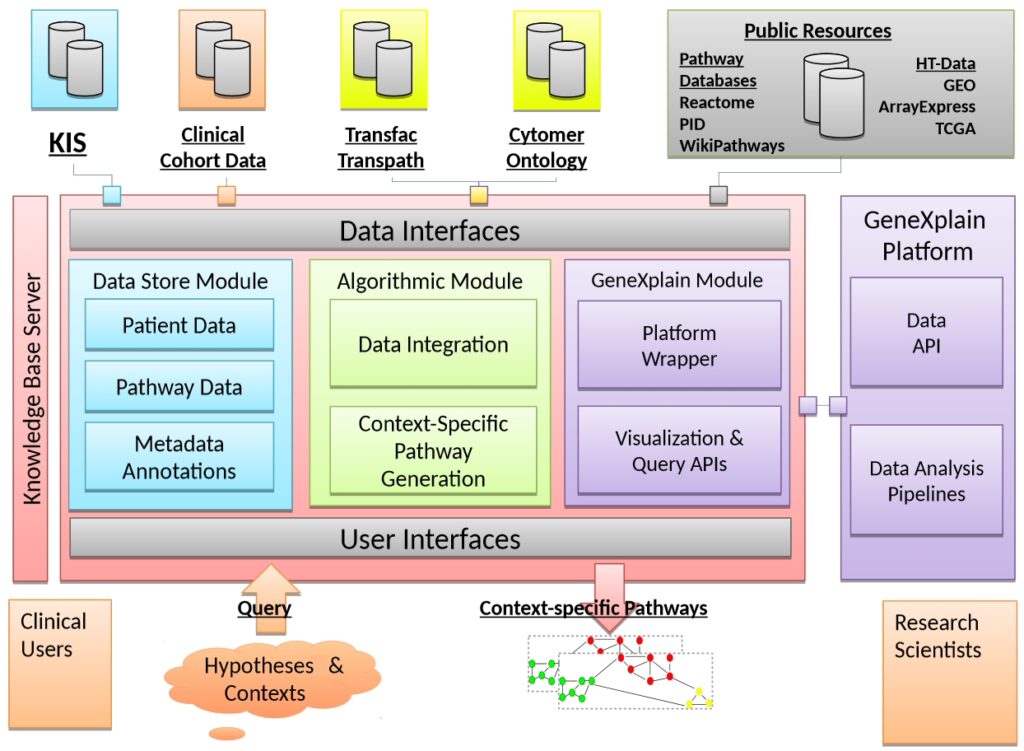
Molecular biomarkers play a role in the diagnosis of cancer. Individualized treatment decisions and specialized drugs warrant the need to broaden the focus from singular biomarkers to pathways. While Omics technologies allow the parallel measurement of many different markers, pathway databases offer vast amounts of knowledge on biological networks. The aim of the MyPathSem project is to present the most relevant, meaningful and interpretable patient-specific pathways to clinicians and researchers. More specifically:
- to link clinical information systems with patient-specific Omics data;
- to generate a tool for data integration and easy access in a clinical environment;
- to collect information from public pathway and literature databases;
- to develop methods to generate context-specific pathways from individual patient data;
- to apply the new tools on data from colorectal and metastatic cancer;
- to test the utility in a clinical research setting; to integrate the tools into the geneXplain platform to make it available to a broader community; and, thus, to reduce the gap between patient centered routine documentation and ontology-driven pathway and gene annotation and establish a seamless data-flow from single patient data to Systems Medicine.
It is geneXplain’s task to contribute a user-friendly computer platform and to provide a prototypic version of an integrated toolbox that can be disseminated among research clinicians for systematic and large-scale validation of the project achievements. GeneXplain will also help disseminating the toolbox through its commercial channels, thereby contributing to the required sustainability of the project.
SysmedIBD

SysmedIBD– Systems medicine of chronic inflammatory bowel disease


A collaborative project funded by the European Union Seventh Framework Programme (FP7/2007-2013) under grant agreement no. 305564, research area FP7-HEALTH-2012-INNOVATION-1, topic HEALTH.2012.2.1.2-2: Systems medicine: Applying systems biology approaches for understanding multifactorial human diseases and their comorbidities.
By applying a systems-biological approach, the project SysmedIBD aims at revealing oscillating biological processes associated with the inflammatory bowel disease (IBD). IBD is a major health problem with severe co-morbidities, requiring life-long treatment. Oscillating processes such as circadian rhythms are well studied and modeled in a number of systems. In the case of IBD as in other inflammatory processes, one particular transcription factor (NF-kappaB) is known to play a key role. Recently, NF-kappaB-associated processes have also been shown to be oscillating. In this project, we will study NF-kappaB oscillation in chronic inflammatory bowel diseases in animal models and patient cohorts with immunosuppressive treatments and controls. The aim is to build an experimentally validated mathematical model of the NF-kappaB oscillation in the gut tissue.
The validated model will enable searching for critical molecular components of the NF-kappaB oscillation and to assess their relevance for the disease in patients.
Gaining control of the oscillation of biological pathways may provide new possibilities to influence biological processes like inflammation. Hence, we will search (assisted by the models and databases developed) for small molecules interfering with the NF-kappaB oscillation and validate selected candidates in experimental systems. Owing to its highly focused system biological approach, the project will generate substantial insights into key mechanisms underlying IBD.
GeneXplain will be involved in the dynamic simulation of the oscillating processed described above, and in the identification of suitable targets and drug candidates, which will then be experimentally validated by the partners.
MIMOmics

Methods for Integrated analysis of Multiple Omics datasets


A collaborative project funded by the European Union Seventh Framework Programme (FP7/2007-2013) under grant agreement no. 305280, research area FP7-HEALTH-2012-INNOVATION-1, topic HEALTH.2012.2.1.1-3: Statistical methods for collection and analysis of –omics data.
MIMOmics aims at developing robust and efficient statistical methods for the integrated analysis of metabolomics, glycomics, proteomic and genomic datasets in large studies.
It was triggered by the repeated observation that the existing methodology for analyzing huge data sets does not match by far the complexity of the biological problem addressed. Expensive and complex data are gathered and being analysed in a rather simple way, thereby missing the opportunity to uncover combinations of predictive and meaningful profiles among the different kinds of omics data. Novel methods should integrate multilevel omics data to bring biological understanding to the next level. Due to the high dimensionality of the datasets the multiple testing burden becomes heavier and consequently the false positive and negative rates increase. Super-meta methods combining multilevel data across populations need to be developed.
Due to the high dimensionality of the datasets the multiple testing burden becomes heavier and consequently the false positive and negative rates increase. Super-meta methods combining multilevel data across populations need to be developed.
GeneXPlain will provide the consortium with the geneXplain platform for data handling and analysis in order to integrate databases of molecular networks with state of the art bioinformatics and systems biology tools. This way, we will support the analysis of pre-existing omics data and apply them for building efficient systems biology models. We will also work on integrating analytical tools developed within the MIMOmics project with already existing systems biology data and knowledge. GeneXplain will be strongly involved in the project’s dissemination activities, thereby particularly considering potential partners from industries.
Mediomics

Mediomics – Systems biology modeling of diseases through integration of multiple “-omics” data


A collaborative Russian-German project funded by the German Federal Ministry of Education and Research (BMBF) and the Russian Fund for Assistance to Small Innovative Enterprises (FASIE).Coordinator:
Dr. Alexander Kel, geneXplain GmbH. The 3 partners are University Regensburg and University Medical Center Göttingen from Germany and biosoft.ru from Russia.Funding period: December 1st, 2013 – August 31st, 2016 (with extension)
Further information:
Project page at the University Medical Center Göttingen
ExITox

ExITox – Explain Inhalation Toxicity


A collaborative project funded by the German Federal Ministry of Education and Research (BMBF) in the funding program “e:ToP – Innovative Toxikologie zur Reduzierung von Tierversuchen”Coordinator:
The project was coordinated by Dr. Sylvia Escher, Fraunhofer Institute for Toxicology and Experimental Medicine (ITEM) in Hannover, Germany. The 6 participants of the consortium are from Hannover, Freiburg, Göttingen, and Wolfenbüttel.
Contract period: November 1st, 2013 – October 31st, 2015, extended until January 31st, 2016
Grant no.: 031 A269 D
Summary:
This project aims at developing an integrated testing strategy (ITS) for the human health risk assessment of repeated dose toxicity after inhalation exposure for the replacement of de novo animal testing. In the pilot phase chemicals with different mode of action will be selected and tested with human precision cut lung slices (PCLS) and human pulmonary cell cultures in order to identify route specific biomarkers. Genome wide transcriptome analyses will be conducted in these models and evaluated using bioinformatics methods. These results will be complemented with data mining results and QSAR predictions. Further, structurally related chemicals will be tested in addition to investigate the possibility of the test system to support read across. The outcome of this pilot project will be a proposal for an integrated testing strategy for respiratory toxicity. Further validation e.g. testing of a broader spectrum of chemicals is foreseen in a follow up project. The proposed ITS, the developed methodologies on data sharing and data integration are not limited to the evaluation of transcriptome data but allow to integrate proteome and metabolome data in a follow up project.
It is geneXplain’s task to analyze transcriptome data of the partners and contribute to the specification of new biomarkers.
Further information:
Homepage of the project at Fraunhofer ITEM
Fraunhofer-Datenbank RepDose
Research Database at Albert Ludwigs University Freiburg (in German)
Publications:
Koschmann, J., Bhar, A., Stegmaier,P., Kel, A. E. and Wingender, E. (2015) “Upstream Analysis”: An integrated promoter-pathway analysis approach to causal interpretation of microarray data. Microarrays 4, 270-286. doi:10.3390/microarrays4020270. Link
Niehof, M., Hildebrandt, T., Danov, O., Arndt, K., Koschmann, J., Dahlmann, F., Hansen, T. and Sewald, K. (2017) RNA isolation from precision-cut lung slices (PCLS) from different species. BMC Res. Notes 10, 121. doi: 10.1186/s13104-017-2447-6. Link
The project continues with ExITox-II.
SYSCOL

Systems Biology of Colorectal Cancer


A collaborative project funded by the European Union Seventh Framework Programme ( FP7/2007-2013 ) under grant agreement no. 258236, research area HEALTH.2010.2.1.2-1, Tackling Human Diseases through Systems Biology Approaches.Coordinator:
The project is coordinated by Prof. Jussi Taipale, Karolinska Institute, Uppsala, Sweden. The 11 participants of the consortium are from 8 European countries, from Israel and the United States.
Contract period: January 1st, 2011 – December 31st, 2015
Contract no. 258236.
Summary:
SYSCOL is a systems biological project that aims at developing a quantitative and comprehensive model of colorectal tumorigenesis, one of the most common cancers in both males and females.
GeneXPlain contributes to the project by developing an integrative tool platform for bioinformatic and systems biological software and databases.
Further information:
Homepage of SYSCOL.
Press release of the Karolinska Institute
Project description on the CORDIS server of the European Commission
Jussi Taipale’s collection of Positional Weight Matrices, the most comprehensive experimental set available to date (Jolma et al., Cell 153: 327-338, 2013; PMID 23332764), has been consistently linked to E. Wingender’s transcription factor classification.
Publications:
Kel, A., Boyarskikh, U., Stegmaier, P., Leskov, L. S., Sokolov, A. V., Yevshin, I., Mandrik, N., Stelmashenko, D., Koschmann, J., Kel-Margoulis, O., Krull, M., Martínez-Cardús, A., Moran, S., Esteller, M., Kolpakov, F., Filipenko, M. and Wingender, E. (2019) Walking pathways with positive feedback loops reveal DNA methylation biomarkers of colorectal cancer. BMC Bioinformatics 20 (Suppl 4), 119. doi: 0.1186/s12859-019-2687-7 Link
Wingender, E., Schoeps, T., Haubrock, M., Dönitz, J. (2015) TFClass: a classification of human transcription factors and their rodent orthologs. Nucleic Acids Res., in press. doi: 10.1093/nar/gku1064. Link
Wingender, E. (2013) Criteria for an updated classification of human transcription factor DNA-binding domains. J. Bioinform. Comput. Biol. 11, 1340007. doi: 10.1142/S0219720013400076. Link
Wingender, E., Schoeps, T., Dönitz, J. (2013) TFClass: An expandable hierarchical classification of human transcription factors. Nucleic Acids Res. 41, D165-D170. doi: 10.1093/nar/gks1123. Link
EPIMETAB
EPIMETAB Epigenetic mechanisms underlying predisposition to metabolic syndrome-associated diseases


A collaborative project funded by the German Federal Ministry of Education and Research (BMBF) in a Joint Transnational Call 2011 for “Integrated Research on Genomics and Pathophysiology of the Metabolic Syndrome and the Diseases arising from it”
Coordinator:
The project is coordinated by Prof. Dr. Pierre Fafournoux, Institut National de la Recherche Agronomique in Saint Genès Champanelle, France. The 4 participants of the consortium are from France (Saint Genès Champanelle and Paris), Spain (Barcelona), and Germany (Wolfenbüttel).
Contract period: April 1st, 2012 – March 31st, 2015
Summary:
The project aims at gaining a better insight on the etiology of different disease conditions such as obesity or type-2 diabetes by investigating the intertwining of genetic, epigenetic and environmental factors. This innovative project is expected to pave the way to new preventive and therapeutic approaches at the individual level and for ameliorating the quality of life at the general population level.
GeneXplain contributes to the project its expertise in promoter analysis and its platform, which will be adapted to the specific needs of the project partners. At the end, a novel integrative view on the regulatory network comprising genetic and epigenetics influences will be provided.
Further information: Project summary (pdf; 8kB)
RESOLVE

A systems biology approach to RESOLVE the molecular pathology of two hallmarks of patients with metabolic syndrome and its co-morbidities; hypertriglyceridemia and low HDL-cholesterol


Through combining basic pre-clinical and clinical research, network analysis and computational modelling, RESOLVE aims at resolving the disturbed dynamics and mechanisms underlying the high triglyceride (HTG) and low high-density lipoprotein cholesterol (HDL-C) phenotype and insulin resistance in patients with metabolic syndrome (MetS) and its associated co-morbidities (cardiovascular disease, CVD; type 2 diabetes, T2DM; non-alcoholic fatty liver disease, NAFLD). RESOLVE will deliver on 6 specific objectives:
- build a computational model for analyzing the kinetics of plasma lipids, lipoproteins and their interactions with glucose metabolism;
- apply the iterative systems biology cycle for calibrating, validating and improving the computational model in dedicated studies in mice;
- build, calibrate and validate the computational model for use in humans;
- analyze – based on model and experimental data – which processes in the murine metabolic network regulatethe physiological response to perturbations in lipid, lipoprotein and glucose metabolism and how these interact;
- analyze – based on model and experimental data – which processes in the human metabolic network regulatethe physiological response to perturbations in lipid, lipoprotein and glucose metabolism and how these interact;
- use the human model to identify network-based drug targets aimed at restoring the metabolic dyslipidemia and glycemic control in patients with MetS and associated comorbidities.
The 60-months project RESOLVE will result into a novel strategy for development of therapeutic strategies for MetS patients with T2DM, NAFLD or CVD.
GeneXplain’s role is to contribute to the general IT infrastructure of the project, to use the company’s technology platform for providing a database of the project-specific data, and to help with training courses for the data management.
GERONTOSHIELD

GERONTOSHIELD – Systems biology-driven approach to unravel and revert the mechanisms responsible for poor immune responses in the elderly


A collaborative project funded by the German Federal Ministry of Education and Research (BMBF) in the funding program “Systems Biology for Health in the Old Age – GerontoSys2”Coordinator:
The project is coordinated by Prof. Dr. Carlos Guzmán, Helmholtz Centre for Infection Research (HZI) in Braunschweig, Germany. The 6 participants of the consortium are from Braunschweig, Jena, Martinsried, Regensburg, Tübingen, and Wolfenbüttel.
- Contract period: February 1st, 2011 – January 31st, 2014
- Grant no.: FKZ0315890B
- Summary:
The strategic objective of the GERONTOSHIELD project is to draw on a Systems Biology-driven approach to understand the molecular processes affected during immunosenescence, in order to derive (i) global mathematical models describing the involved processes, (ii) a holistic model for flu infections in young and elderly, and (iii) strategies to overcome these processes to establish immune interventions tailored for the elderly.
Further information:
Homepage of GERONTOSHIELD
Press release of the Helmholtz Centre for Infection Research (HZI; in German)
Press release of the Helmholtz Centre for Infection Research (HZI; in English)
Publications:
Wingender, E. (2013) Criteria for an updated classification of human transcription factor DNA-binding domains. J. Bioinform. Comput. Biol. 11, 1340007. doi: 10.1142/S0219720013400076. Link
Wingender, E., Schoeps, T., Dönitz, J. (2013) TFClass: An expandable hierarchical classification of human transcription factors. Nucleic Acids Res. 41, D165-D170. doi: 10.1093/nar/gks1123. Link
TEMPUS

TEMPUS project “Rehaut”

A collaborative educational & training project financially supported by TEMPUS program of the European Union, phase TEMPUS IV, under grant agreement no. 511426-TEMPUS-1-2010-1-RU-TEMPUS-JPCR.Coordinator:
The project is coordinated by Prof. Alexander Kuznetsov, D. Mendeleyev University of Chemical Technology of Russia, Moscow. The participants of the consortium are from 3 EU member states, from Russia and Belorus.
Contract period: October 15th, 2010 – October 14th, 2013
Summary:
The project aims at harmonizing, standardizing and developing further the European BSc and MSc biotechnology curricula and their applicability to biotechnology education in Belorus and Russia. It is geneXplain’s contribution to reinforce the industrial view in this project, in particular with regard to the inclusion of the methods of computational biology.
Further information:News about the project from the National TEMPUS Office of Belorus.
Rehaut Tempus project page (in Russian)
