Free webseminars
Welcome to geneXplain’s free webseminars!
Upcoming webseminars:
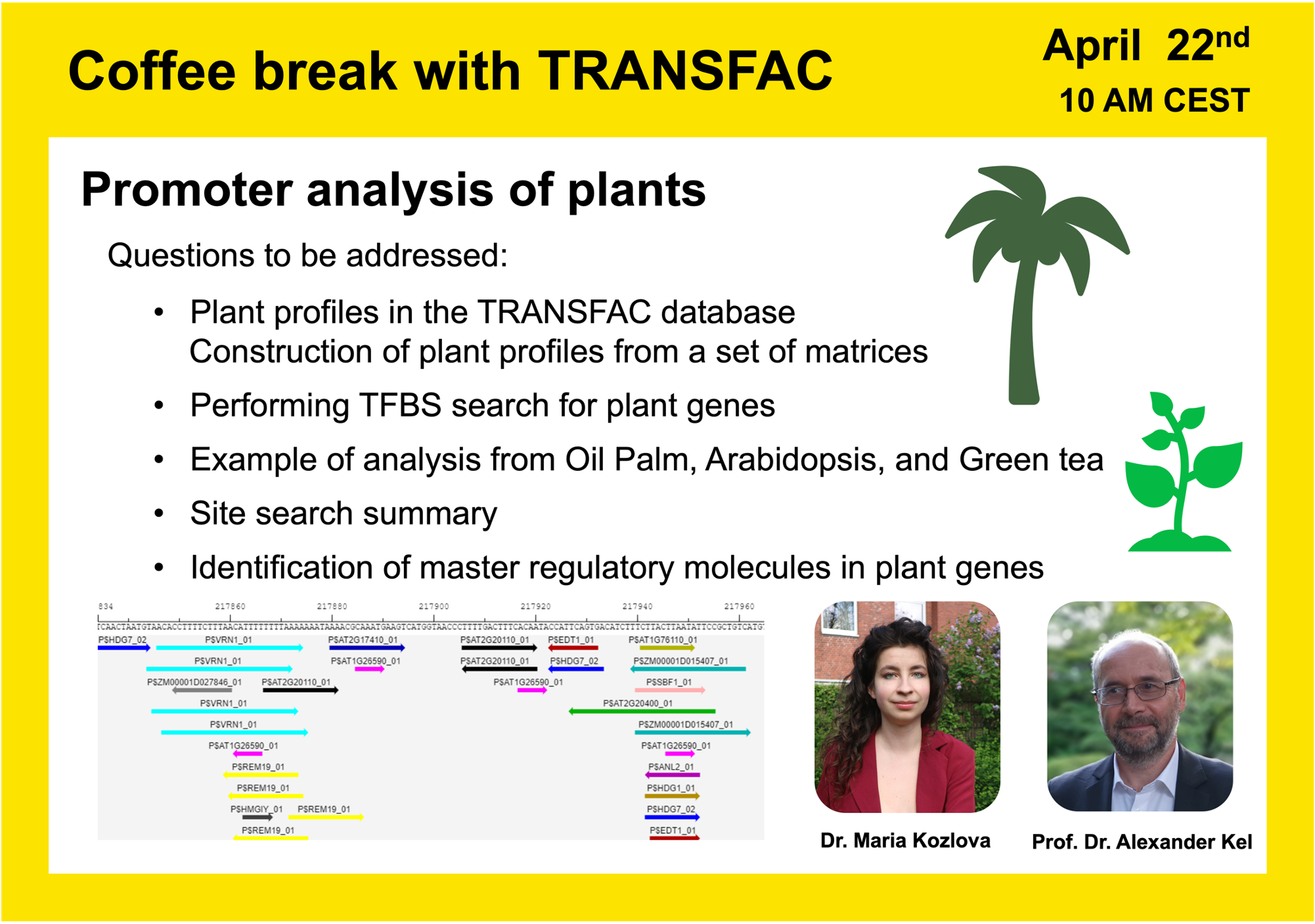
The next Coffee break with TRANSFAC will be will be held on April 22nd at 10 AM CET
Fill in this form to receive the event joining link
Promoter analysis of plants
In this webinar Dr. Alexander Kel, the CEO and CSO of geneXplain GmbH, together with the biologist from Osnabrück University Dr. Maria Kozlova, will apply TRANSFAC to study different Plant Promoters. The following main questions will be in the focus of this seminar:
- What is the prerequisite to integrate a plant genome?
- What plant profiles are there in TRANSFAC?
- How to analyze different types of plant data inside the geneXplain platform?
- How to build your own pipeline?
Stay tuned to our news for information on our next planned events.
Welcome to view our recent webseminars:
- Coffee breaks with TRANSFAC
- Young cerebrospinal fluid restores oligodendrogenesis and memory in aged mice via Fgf17
- Delineating the role of master transcriptional regulator mediated signalling in disease progression and treatment response in gastrointestinal disorders
- World Cancer Research Day at geneXplain:
– Cancer as regulatory disease
– Genome Enhancer: from raw omics data to personalised drug targets and treatments
– TRANSFAC 2.0: the fresh look at the gold standard of gene regulation studies
- A computationally aided strategy to recover gene activity profiles from post-mortem HIV+ brain specimens
- Decoding non-coding genome: How and why mutations in TF binding sites may lead to cancer or cytokine storm
- In silico evaluation of the pharmacological potential of natural compounds
- TRANSFAC® database and its applications
- Transcription factors and their classification
- Regulatory SNPs in TRANSFAC® sites. Severe COVID-19 patients versus non-symptomatic.
- COVID-19 Challenge: Known unknowns and unknown unknowns in biological activity spectra of the current and emerging pharmaceutical agents.
- COVID-19 Challenge: In silico approach to finding potential anti-SARS-CoV-2 agents among one billion molecules
- COVID-19 – online discovery of prospective drug targets and personalized treatments with Genome Enhancer
- COVID-19 – online discovery of prospective drug targets and treatments with Genome Enhancer
Young cerebrospinal fluid restores oligodendrogenesis and memory in aged mice via Fgf17
This webseminar was held on July 12th, 2022
The webseminar was given by our invited speaker Dr. Tal Iram, the post-doctorate fellow at the Department of Neurology and Neurological Sciences, Stanford University School of Medicine, Stanford, CA, USA
Further info
Dr. Tal Iram is the corresponding author of the eponymous Nature paper “Young CSF restores oligodendrogenesis and memory in aged mice via Fgf17”, which was widely shared on the news worldwide due to the exciting discovery of mouse brain cell rejuvenation by using the cerebrospinal fluid (CSF) of young mice.
Dr. Tal Iram is the post-doctorate fellow at the Department of Neurology and Neurological Sciences, Stanford University School of Medicine, Stanford, CA, USA. She received her PhD in Neuroscience from Tel Aviv University in Israel in 2017. Shortly after, she began her postdoc in the Wyss-Coray lab at Stanford where she developed a novel rejuvenation paradigm by transferring cerebrospinal fluid (CSF) from young to aged mice. This led her to study oligodendrocyte rejuvenation in the aging hippocampus in the context of cognitive aging. She identified that Fgf17, a CSF growth factor that decreases with age, is sufficient for oligodendrocyte rejuvenation and for improvements in cognitive function. The results of respective study were recently published in this Nature paper.
It is our honor to outline that the associated work has made extensive use of the TRANSFAC database maintained and distributed worldwide by geneXplain GmbH.
You are welcome to watch the video record of this webseminar here.
Delineating the role of master transcriptional regulator mediated signalling in disease progression and treatment response in gastrointestinal disorders
This webseminar was held on May 31st, 2022
The webseminar was given by our invited speaker Dr. Sudipto Das, Lead of the Epigenetics of Gastrointestinal diseases research group, School of Pharmacy & Biomolecular Sciences, Royal College of Surgeons in Ireland, (RCSI) Dublin, Ireland
Webseminar brief summary
Gene expression changes underpinning disease development are orchestrated by a milieu of complex regulatory processes. This talk showcased recent work that demonstrates the role of master transcriptional regulator mediated signalling that underlines differences in anti-tumour immunity between patients of African and European ancestry with colorectal cancer. Furthermore, it was shown how novel epigenetic targeting agents mediate global transcriptional alterations through master regulators and transcription factors in colorectal cancer. Finally, the talk highlighted an ongoing study that rationalises further mechanistic interrogation of these master regulators as potential therapeutic targets to improve response to biologic treatment in Ulcerative colitis.
You are welcome to watch the video record of this webseminar here.
A computationally aided strategy to recover gene activity profiles from post-mortem HIV+ brain specimens
This webseminar was held on May 19th, 2021
The webseminar was given by our invited speaker Dr. Cecilia Marcondes, Associate Professor at San Diego Biomedical Research Institute
Webseminar brief summary
Dr. Marcondes talked about her group’s attempt to overcome a challenge in the research of human tissues, where post-mortem intervals may impair the retrieval of quality information from the transcriptome.
Using epigenetic marks and computational analytical strategies, her group has successfully identified gene networks associated with critical biological processes in post-mortem brains from HIV+ cases, using resources from the National NeuroAIDS Tissue Consortium (NNTC). The combination of systems biology and visualization tools, including IPA, DAVID Bioinformatics Resources, Cytoscape, GeneMania, TRANSFAC FMatch, and iRegulon, led to the identification of genomic regions functionally annotated to inflammation, anti-viral response, and to synaptic pathways with the ability to distinguish individuals with history of substance use disorder in the context of HIV infection. This work demonstrates how creative strategies and computer aided analysis can help increase the power of association studies using valuable post-mortem human tissue.
You are welcome to watch the video record of this webseminar here.
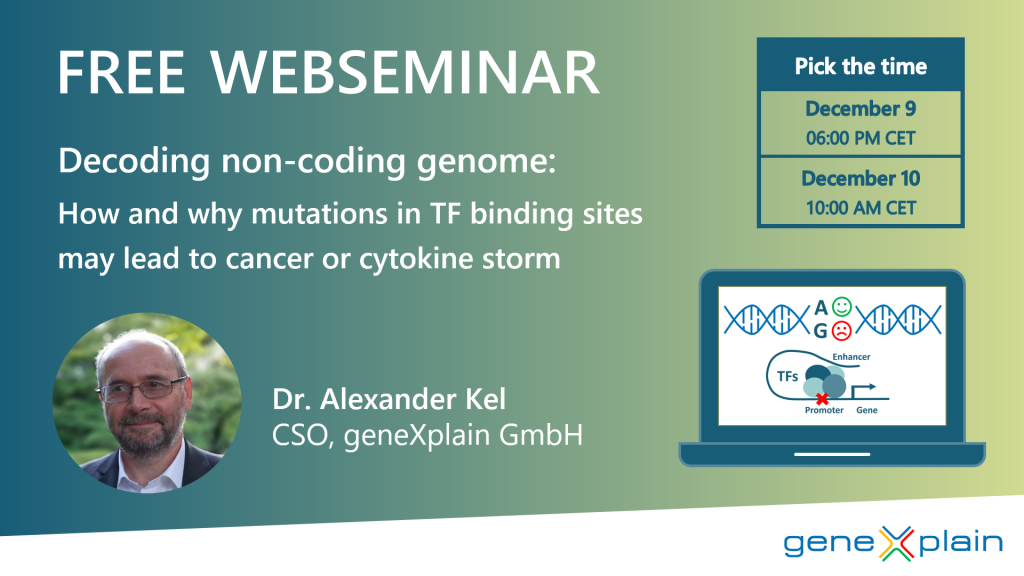
Decoding non-coding genome: How and why mutations in TF binding sites may lead to cancer or cytokine storm
This webseminar was held on December 9th and 10th, 2020.
The webseminar was given by geneXplain’s CSO Dr. Alexander Kel.
Webseminar brief summary
In this webseminar Dr. Alexander Kel presented our concept of the main principles of regulatory code. He began the lecture by discussing the puzzle of how non-coding genomic regions can regulate activity of thousands of genes in millions of different cellular conditions and how this regulation can go wrong in case of diseases, such as cancer and various forms of inflammatory diseases. Composite regulatory elements and modules that consist of several TF binding sites became the focus of discussion, giving a clue to the understanding the gene regulatory code. “Fuzzy puzzle” – is an analogy that was used for such coding principle. Next, Dr. Kel presented the ideas about possible emerging of such fuzzy puzzle as a result of resolving the major limitations of evolution of multicellular organisms during the latest steps of evolution. Finally, results of computational analysis of non-coding genomic regions were presented, describing how AI-driven algorithms can detect cancer-specific and inflammation-specific genomic and epigenomic alterations in composite regulatory modules helping to reveal novel mechanisms of disease progression leading to new avenues of research and ultimately to better diagnostic tests and personalized therapies.
You are welcome to watch the video record of this webseminar here.
Chinese visitors are welcome to view the recording here
观看不了YouTube的用户,可以到 这里 观看网络研讨会视频
In silico evaluation of the pharmacological potential of natural compounds
This webseminar was held on November 18th, 2020, at 6 PM CET and was then repeated due to multiple requests on November 24th, 2020, at 10 AM CET.
The webseminar was given by our invited speaker Prof. Vladimir Poroikov from the Institute of Biomedical Chemistry (IBMC), Moscow, Russia.
Webseminar brief summary
Natural products represent the auspicious source of medicines with improved safety and efficacy. In addition to their inherent biological activities provided as a “Gift of Mother Nature,” they exhibit substantial structural diversity that creates excellent opportunities to discover new pharmaceutical agents.
Medicinal plants are used in the traditional medicines of many countries. For example, Traditional Indian Medicine Ayurveda is one of the world’s oldest medical sciences, with a history that goes back more than 5,000 years. The knowledge of Ayurveda has at various times had an impact on many branches of medicine, and it continues to retain its prominent position nowadays, being officially recognized by the World Health Organization (WHO) and enjoying great popularity in the US, Germany, Italy, and the Netherlands.
The existing databases on natural products used in traditional medicines do not contain exhaustive information about natural products’ pharmacological potential.
At the webseminar Prof. Vladimir Poroikov demonstrated how new medical indications for medicinal plants’ phytoconstituents may be identified by applying bio- and chemoinformatics methods.
Several case studies were considered, including:
- Revealing medicinal plants useful for the comprehensive management of epilepsy and associated comorbidities.
- Repositioning of Achyranthes Aspera as an antidepressant agent.
- Discovery of the molecular mechanism of N-acetyl aspartate anti-aging action.
You are welcome to watch the video record of this webseminar here.
Chinese visitors are welcome to view the recording here
观看不了YouTube的用户,可以到 这里 观看网络研讨会视频
TRANSFAC® database and its applications
This webseminar was held on October 21st, 2020.
Webseminar brief summary:
The TRANSFAC® database is the gold standard in the area of transcriptional regulation. Dating back to a very early compilation, this database of eukaryotic transcription factors, their genomic binding sites and DNA-binding profiles has been carefully maintained and manually curated for over 30 years.
At the webseminar the founder and initiator of TRANSFAC® database Prof. Dr. Edgar Wingender, CEO of geneXplain GmbH, guided us though the historical origins, basic concepts and current status of gene regulation aspects in the context of TRANSFAC® database. Special emphasis was given to the recent developments of TRANSFAC® database, which fully address the latest needs of gene regulation studies.
The second part of the webseminar was presented by Dr. Volker Matys, who has been working on TRANSFAC® for more than twenty years and is currently in charge of customer support for TRANSFAC® at geneXplain GmbH. Dr. Volker Matys demonstrated TRANSFAC® applications for various biological research needs with focus on analysis of genomic sequences for potential transcription factor binding sites with the tools Match™ and FMatch.
Transcription factors and their classification
This webseminar was held on September 16th, 2020, at 6 PM CEST
Webseminar brief summary:
Transcription factors (TFs) are proteins that regulate the gene activity program of a cell. They direct the RNA polymerase to the correct start site of transcription in a cell- and stage-specific way, frequently responding to extracellular stimuli.
Most TFs bind to DNA in a somewhat relaxed sequence-specific way, thereby decoding the regulatory signals of the genome. Comparative analysis of their DNA-binding domains (DBDs) can be used (1) to establish a classification system for this important group of proteins; (2) to find a clue to decipher the DNA-protein recognition code; (3) to elucidate the evolutionary history of TFs; (4) to derive computational models for these DBDs that help identify orthologous TFs encoded by newly sequenced genomes and, thus, establish TF catalogs for these species.
This webseminar addressed the basic mechanism of transcription and features of transcription factors in eukaryotes, in particular their mode of recognizing regulatory DNA elements. The principles of a TF classification were outlined, their implementation as TFClass was depicted.
When applying these classification principles to different species, it has been revealed that TFs of some classes are instances of very old principles, whereas others are subject to very recent dynamic evolution.
You are welcome to watch the video record of this webseminar here.
Chinese visitors are welcome to view the recording here
观看不了YouTube的用户,可以到 这里 观看网络研讨会视频
Regulatory SNPs in TRANSFAC® sites. Severe COVID-19 patients versus non-symptomatic.
This webseminar was held on August 5th, 2020
The webseminar was given by geneXplain’s CSO Dr. Alexander Kel.
Webseminar brief summary:
More then 90% of disease-linked SNPs are non-coding. So, they are located outside of the protein coding regions of genome and often mark the genomic regions that regulate genes whose aberrant expression leads to dysfunction of signal transduction or metabolic pathways leading to the elevated disease susceptibility or severity. Such regulatory SNPs are difficult to interpret since the regulatory code is still purely understood. Regulatory SNPs are often associated with DNA sequence variations that effect transcription factor bindings sites (TFBSs), which leads to the change in regulation of the target genes. We used TRANSFAC® database and the geneXplain® platform with the automatic omics analysis tool Genome Enhancer to analyze TFBS of SNPs revealed in GWAS study in the COVID-19 Host Genetics Initiative, which is a global initiative to elucidate the role of host genetic factors in susceptibility and severity of the SARS-CoV-2 virus pandemic. Eur. J. Hum. Genet. 28, 715–718 (2020). https://doi.org/10.1038/s41431-020-0636-6.
We revealed TFBSs (for such TFs as FOXO1, TCF3, E2F1, TP53) strongly associated with SNPs linked with the severity of COVID-19 disease. Upstream analysis of these TFs in the context of the reveled SNPs gave us a clue about signal transduction pathways associated with the potential predisposition to the severe COVID-19 disease.
You are welcome to watch the video record of this webseminar here.
Chinese visitors are welcome to view the recording here
观看不了YouTube的用户,可以到 这里 观看网络研讨会视频
COVID-19 Challenge: Known unknowns and unknown unknowns in biological activity spectra of the current and emerging pharmaceutical agents
This video is a record of geneXplain’s live webseminar ‘COVID-19 Challenge: Known unknowns and unknown unknowns in biological activity spectra of the current and emerging pharmaceutical agents’ which was held on June 24th, 2020.
This webseminar was given by our invited speaker, Prof. Vladimir Poroikov from the Institute of Biomedical Chemistry (IBMC), Moscow, Russia.
At the webseminar Prof. Vladimir Poroikov showed how to apply human intelligence and machine learning methods to in silico analysis of biological activity profiles of the launched and investigational drugs currently proposed for the pharmacotherapy of multifaceted clinical manifestations of SARS-CoV-2 infection.
Particular emphasis was placed on PharmaExpert software that analyzes the biological activity spectra predicted by PASS taking into account the relationships between different biological activities, and identifies the pleiotropic effects of drug combinations and preparations of medicinal plants.
You are welcome to watch the video record of this webseminar here.
Chinese visitors are welcome to view the recording here
观看不了YouTube的用户,可以到 这里 观看网络研讨会视频
COVID-19 Challenge: In silico approach to finding potential anti-SARS-CoV-2 agents among one billion molecules
This video is a record of geneXplain’s live webseminar ‘COVID-19 Challenge: In silico approach to finding potential anti-SARS-CoV-2 agents among one billion molecules’ which was held on May 13th, 2020.
This webseminar was given by our invited speaker, Prof. Vladimir Poroikov from the Institute of Biomedical Chemistry (IBMC), Moscow, Russia.
At the webseminar Prof. Vladimir Poroikov showed how to apply artificial intelligence and machine learning methods to in silico screening of hits with the desired biological activity in chemical libraries of synthesized or virtual molecules.
Particular emphasis was placed on PASS (Prediction of Activity Spectra for Substances), one of the earliest and most widely used software for the elucidation of hidden pharmacological potential of drug-like molecules.
You are welcome to watch the video record of this webseminar here.
Chinese visitors are welcome to view the recording here
观看不了YouTube的用户,可以到 这里 观看网络研讨会视频
COVID-19 – online discovery of prospective drug targets and personalized treatments with Genome Enhancer
This video is a record of geneXplain’s live webseminar ‘COVID-19 – online discovery of prospective drug targets and personalized treatments with Genome Enhancer’ which was held on April 29th, 2020.
In this video geneXplain’s CSO Dr. Alexander Kel is performing the analysis of personalized transcriptomics data coming from bronchoalveolar lavage of certain COVID-19 patients using the Genome Enhancer software.
You are welcome to watch the video record of this webseminar here.
Chinese visitors are welcome to view the recording here
观看不了YouTube的用户,可以到 这里 观看网络研讨会视频
COVID-19 – online discovery of prospective drug targets and treatments with Genome Enhancer
This video is a record of geneXplain’s live webseminar ‘COVID-19 – online discovery of prospective drug targets and treatments with Genome Enhancer’ which was held on April 7th, 2020.
In this video geneXplain’s CSO Dr. Alexander Kel is performing the integrated analysis of transcriptomics and proteomics datasets of COVID-19 patient data in search for prospective drug targets and treatments using the Genome Enhancer software.
You are welcome to watch the video record of this webseminar here.
Chinese visitors are welcome to view the recording here
观看不了YouTube的用户,可以到 这里 观看网络研讨会视频
Thank you for your interest in our webseminars!
If you have any suggestions for new topics of our webseminars, please let us know via info@genexplain.com, and maybe your topic will be selected for one of the next webseminars!